Research groups in the Regensburg Center for Biochemistry
 |
Patrick Babinger | Functional Assignment of Uncharacterized EnzymesUsing a combination of computational biology, biochemistry and genetics, we want to provide insights into the evolution of novel enzymatic activities and to reveal previously unknown biosynthetic pathways. Our current research focuses on the biosynthesis of ether lipids, which are well known in Archaea. We elucidate their previously unknown occurrence in Bacteria, study novel ether lipid modifications and reconstruct the evolution of the involved enzymes. |
 |
 |
Astrid Bruckmann | Mass spectrometry/ProteomicsOur group aims at providing state-of-the-art mass spectrometric methods which are tailored to the project specific requirements of our collaboration partners within the RCB and beyond. We are particularly focusing our interest on quantitative proteomics, interactomics (RNA-and protein proximity labeling workflows), cross-link (XL)-MS methods, analysis of post-translational modifications as well as nascent proteomics. |
 |
 |
Neva Caliskan | RIBOx ; Research in RNA Interactions, biochemistry and organizationProf. Neva Caliskan’s research group investigates RNA-mediated mechanisms that regulate gene expression, particularly in the contexts of infection, cellular differentiation, and stress responses. To achieve this, the team utilizes fluorescence-based and force-based single-molecule techniques, along with ensemble kinetic analyses, to reveal detailed mechanistic insights and dynamic interactions between RNA and proteins with high spatiotemporal resolution.. |
 |
 |
Christoph Engel | Structural BiochemistryTo understand the molecular basis of transcription by eukaryotic RNA polymerases we study their structures in vitro. For this purpose, we use state-of-the-art Structural Biology tools, such as cryo-Electron Microscopy and X-ray Crystallography, and develop tailored functional assays to elucidate structure-function relations. |
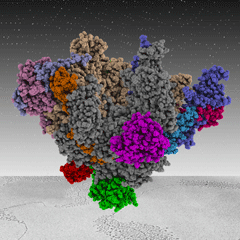 |
 |
Klaus Grasser | Chromatin, Gene Expression and DevelopmentIn focus of our research is the transcriptional elongation by RNA polymerase II. Of particular interest are factors that regulate the efficiency of transcript elongation in the context of chromatin. In addition, the co-transcriptional recruitment of proteins is analysed that ultimately mediate the export of mature mRNAs from the cell nucleus. We study the role of these processes in the control of plant development and the response to environmental stress. |
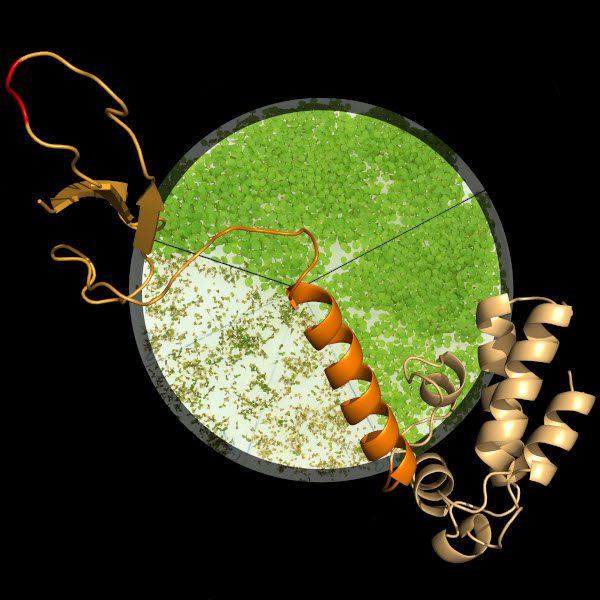 |
 |
Joachim Griesenbeck | rDNA ChromatinOur aim is to increase the understanding of how eukaryotes regulate transcription of their genome in the cell nucleus. We focus on the specific role of chromatin structure within this essential process. Our favorite model loci are the ribosomal RNA genes in the yeast S. cerevisiae, which we investigate by using biochemistry, cell biology, genetics, molecular biology, and structural biology. |
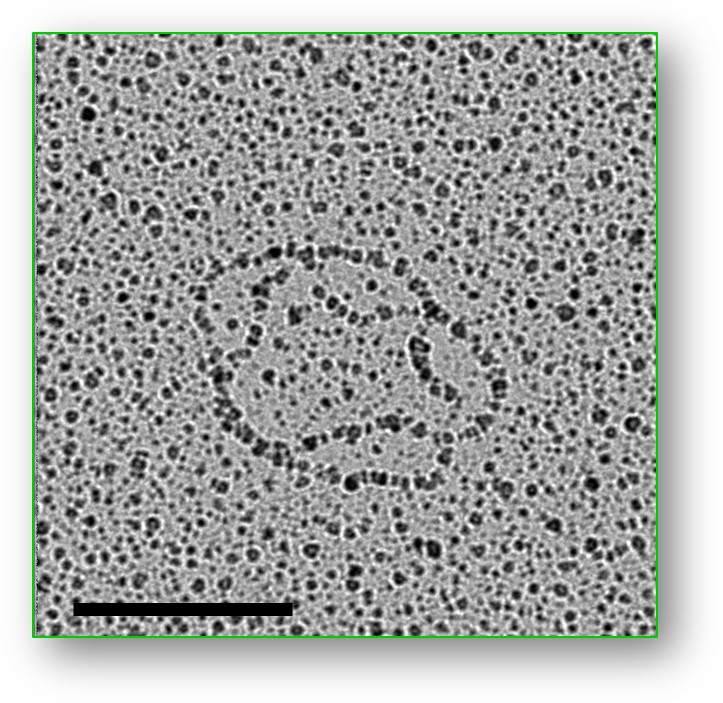 |
 |
Dina Grohmann | Single Molecule BiochemistryWe integrate genetic, biochemical and biophysical methods to investigate processes along the gene expression pathway including the regulation of gene expression at the transcriptional (archaeal and eukaryotic transcription machinery) and posttranscriptional level (RNA processing, RNA-guided interference and archaeal Sm like proteins) as well as prokaryotic defence system (CRISPR-Cas, prokaroytic Argonaute). We are especially interested to understand molecular heterogeneity and employ single-molecule techniques (esp. single-molecule FRET and Nanopore sequencing) to unravel conformational states of molecular machinieries or gain insights heterogeneous states of the transcriptome.Being home of the Archaea Centre Regensburg, we are particularly interested in the third domain of life, the Archaea. |
 |
 |
Winfried Hausner | Gene Regulation Networks in Pyrococcus FuriosusThe genome of Pyrococcus furiosus encodes 85 putative transcription factors but the function of most of these proteins and the corresponding gene regulatory networks are unknown. To unravel the function of some of these proteins we combine traditional biochemical in vitro techniques to identify binding sequences and transcription profiles of individual genes with global approaches like ChiP-seq or RNA sequencing to reveal gene regulatory networks. Our latest project deals with CopR, a transcription regulator, which is necessary to survive at elevated copper concentrations. |
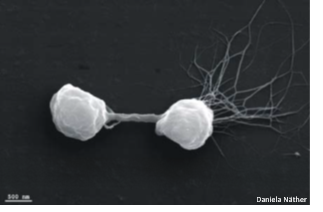 |
 |
Andrea Hupfeld (née Kneuttinger) | Engineering of Photocontrol in EnzymesPhotocontrol of proteins is a rapidly growing research field that facilitates the spatio-temporal regulation of biological activities with light. We engineer artificial enzymes with an internal activity switch by incorporating light-sensitive unnatural amino acids. Our research focuses on continuously developing these switches by combining chemical synthesis and directed evolution and on designing and applying specific photocontrol systems for fundamental research, biotherapy and biocatalysis. |
 |
 |
Aline Koch | Exogenous RNA applications in plantsMy group focuses on non-coding and coding RNAs and exploits the molecular mechanisms (transcription and translation, DNA and histone modifications, mRNA degradation) regulated by these RNAs for plant applications. We have long-standing expertise in RNAi-based plant protection using both GM and non-GM strategies and have pioneered the spray-induced gene silencing strategy to control plant diseases caused by fungal pathogens. Our current goal is to identify the molecular mechanisms (uptake, processing, transport) underlying RNA-based technologies that are essential for their successful ''lab to field'' transfer. To achieve this, we are integrating our findings in a highly interdisciplinary framework. |
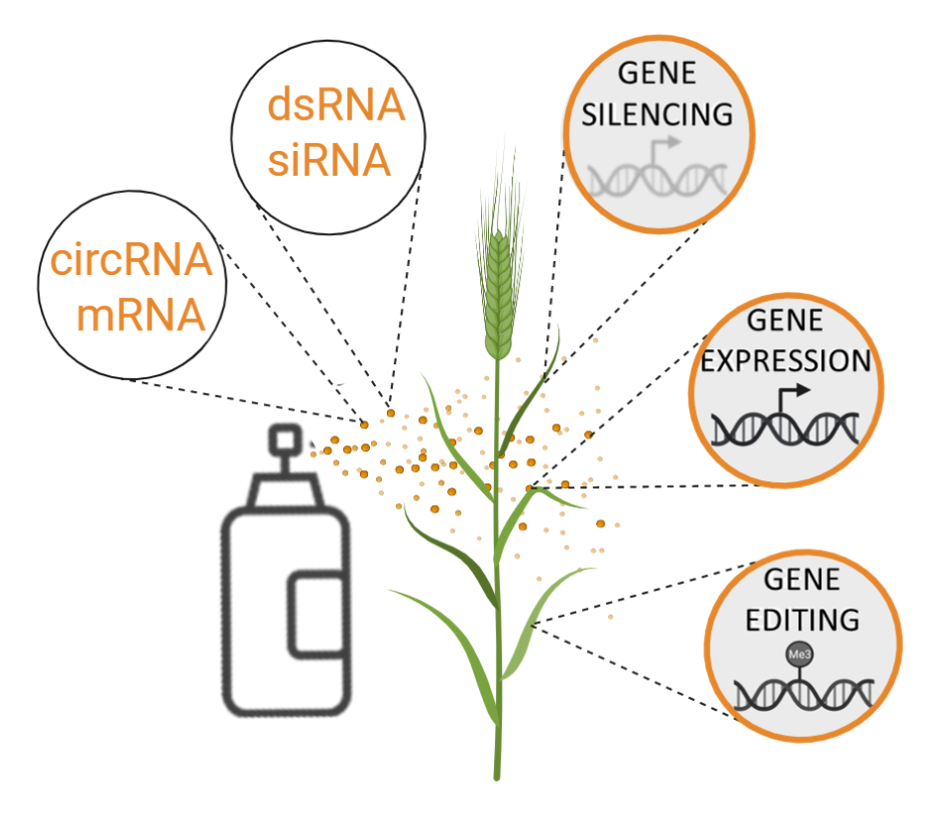 |
 |
Gernot Längst | Chromatin Dynamics and Nuclear ArchitectureThe packaging of eukaryotic DNA into chromatin presents an overriding regulatory layer to all DNA dependent processes, like transcription or replication. We study the role of chromatin dynamics, genome packaging, and genome wide chromatin structures in human cells, the Malaria causing parasite Plasmodium falciparum, the Adenovirus, and the Corona Virus. We employ cell biological, biochemical, biophysical and genomic high-throughput methods to unravel the mechanisms and dynamic changes of genome packaging. Furthermore, we address to role of non-coding RNA in chromatin, showing that these can bind sequence-specific to DNA via triple-helices, regulating chromatin density and the activity of the genome. |
 |
 |
Gunter Meister | Gene Regulation by microRNAsOur research is focused on the investigation of non-coding RNA pathways in mammalian cells. We focus on Post-translational regulation of small RNA pathways, Profiling of non-coding RNAs in cancer, Regulation of gene expression by RNA-binding proteins and Investigation of RNA-specific base modification pathways. |
 |
 |
Nina Ripin | RNP granulesOur research group focuses on understanding the role of RNA, RNA condensation or aggregation and RNA Chaperones in Ribonucleoprotein (RNP) granule formation, disassembly and function in mammalian cells. We use a combination of cell & molecular biology, imaging and biochemical techniques. |
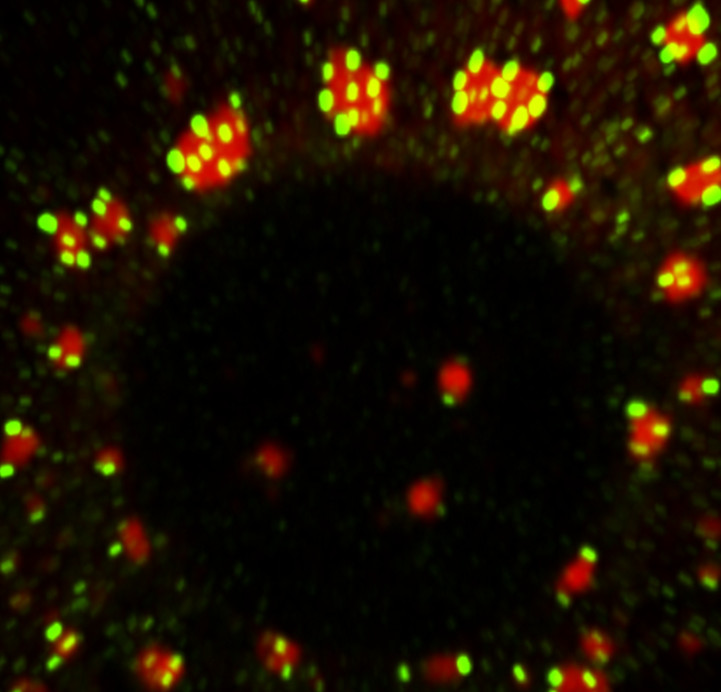 |
 |
Till Rudack | Structural BioinformaticsOur goal is to gain scale-bridging, time-resolved atomic insights into the structure and dynamics of molecular machines of biomedical or biotechnological importance. The derived hypotheses about molecular mechanisms are used, for example, to improve the understanding of disease, to foster drug development, to improve therapeutic and diagnostic methods, or for targeted protein design. To achieve discoveries, we develop and use computational strategies that combine biomolecular simulation algorithms, quantum chemical calculations, and artificial intelligence with data from biophysical and biochemical experiments. |
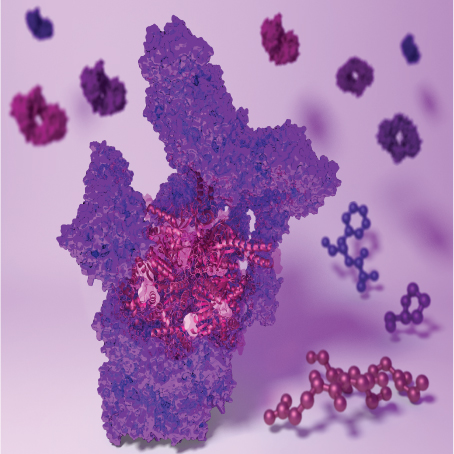 |
 |
Wolfgang Seufert | Regulation of the Cell Division CycleWe are interested in regulatory pathways that organize the division cycle of eukaryotic cells and mainly use budding yeast as a model organism. Specifically, we study how cells complete mitosis and how mRNA translation influences cell cycle entry. In this work, we combine various genetic and biochemical methods and employ flow cytometry and live cell imaging. |
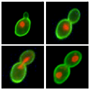 |
 |
Remco Sprangers | Biomolecular NMR SpectroscopyThe primary scientific goal of our research is to understand the relationship between protein motions and protein function. This is especially relevant for enzymes that have to undergo structural rearrangements to perform biological tasks. Many of the projects in the lab we focus on understanding the mechanism behind the bio-molecular complexes that play a role in the degradation of mRNA. To achieve our goals we exploit and develop high resolution NMR methods. |
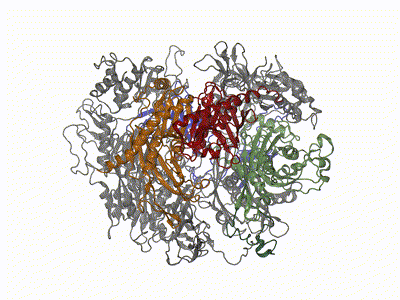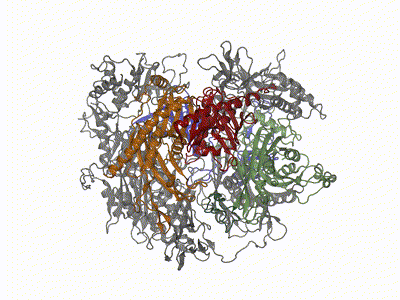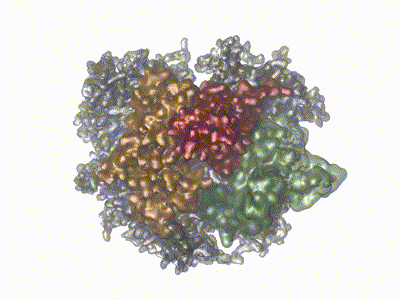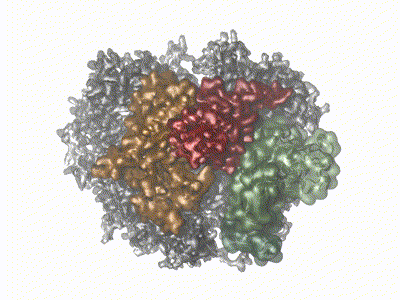 |
 |
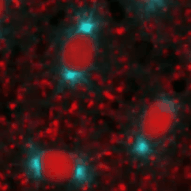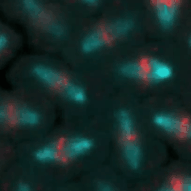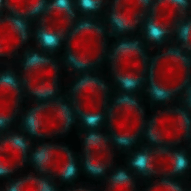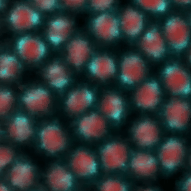
Frank Sprenger | Cell Cycle Control in DrosophilaRegulated proteolysis is a major mechanism of cell cycle progression and cell proliferation. The timed degradation of critical cell cycle regulatory proteins provides tight control and directionality of the cell cycle. Protein destined for degradation are marked by ubiquitin chains that are attached by ubiquitin ligases. We are interested in the regulation of three major ubiquitin ligases, the APC/C (Anaphase promoting complex/cyclosome), SCF (Skp/Cullin/Fbox) und CRL4 (Cullin ring ligase 4) and would like to identify their critical targets. We are using Drosophila as a model system that has the same cell cycle circuitry as mammalians but fewer cell cycle regulatory proteins to unravel the mechanisms of the cell cycle. |
 |
 |
Reinhard Sterner | Enzyme mechanisms, evolution, and engineeringEnzymes are sophisticated proteins that catalyze cellular reactions with high efficiency. We computationally and experimentally reconstruct the molecular evolution of enzymes, use rational design and directed evolution to generate enzymes with a tailored combination of activity and stability, design enzymes that can be controlled by light, and characterize allosteric interactions within multi-enzyme complexes. |
 |
 |
Ander Stiel | Protein Engineering for Superresolution MicroscopyImaging is our window into the processes of life. Advanced imaging techniques rely on an interplay between hardware and innovative imaging agents such as genetically encoded tools, for example, photo-switching proteins. The Stiel lab uses protein engineering techniques to develop such genetically encoded tools for optical imaging techniques such as super resolution microscopy, conventional fluorescence imaging and photo-/optoacoustics imaging. The lab utilises both directed evolution and screening based approaches as well as rational structure driven engineering techniques to develop the proteins. This work is flanked by spectroscopy and structural analysis to understand the molecular determinants of the photophysical properties. |
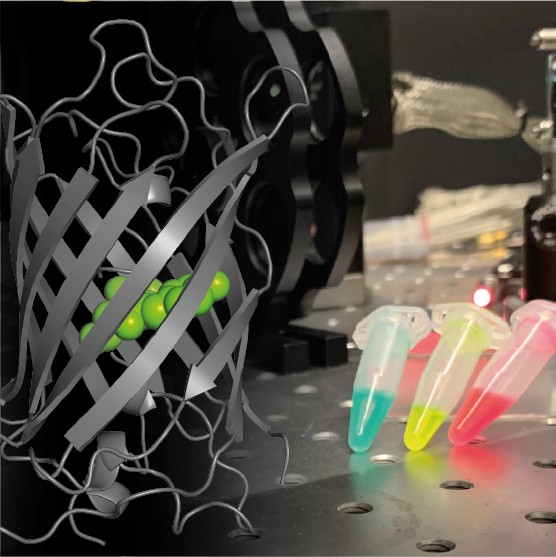 |
 |
Herbert Tschochner | rRNA SynthesisThe main objective of our research is the complex process of eukaryotic ribosome biogenesis. Ribosome synthesis involves transcription of ribosomal RNA genes, processing and assembly of ribosomal precursor molecules in the nucleolus and the nucleus, their transport to the cytoplasm and final maturation. Our special interest is to understand how the rRNA synthesizing machinery is regulated and how it communicates with rRNA processing and ribosome assembly. |
 |
 |
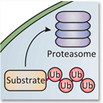
Silke Wiesner | Ubiquitin-dependent Cell SignalingUbiquitin (Ub) and Ub-like (UBL) modifiers are a small family of proteins that regulate virtually all signal transduction pathways in eukaryotes. Aberrant activity of the Ub / UBL modification machineries gives rise to severe cellular dysfunctions that cause numerous human diseases including cancer. We seek to understand how Ub / UBL modification enzymes function on an atomic level, how their dynamics and activities are controlled and how Ub / UBL modification regulates cellular behavior. To this end, we combine structural studies using NMR spectroscopy and X-ray crystallography with biochemical methods. |
 |
 |
Christine Ziegler | Structure and Function of Membrane ProteinsWe focus on the structure and function of membrane proteins and macromolecular complexes, which are involved in stress-regulated and stress-activated transport processes across the membrane. |
 |
Alumni
 |
Markus Jeschek | Synthetic MicrobiologyThe overarching goal of our research is the construction of synthetic bacteria that facilitate sustainable bioproduction as an alternative for petrochemistry. Specifically, we develop molecular tools, experimental techniques and computational models, which allow us to rationalize and streamline the underlying engineering process. To achieve this, we capitalize a combination of high-throughput experimentation, next-generation sequencing and data-driven modeling to improve our understanding of fundamental biological processes such as gene regulation on all levels of the central dogma. |
 |
 |
Sebastien Ferreira-Cerca | Cellular Biochemistry of MicroorganismsOur research aims to obtain functional insights into conserved and specific molecular principles of spatiotemporal organization, regulation, and dynamics of RNA metabolism in microorganisms. To this end, we study the ribosome life cycle as a model system, and apply genetic, cellular, molecular, and biochemical approaches in different model organisms, primarily archaea but also bacteria and yeast. |
 |
 |
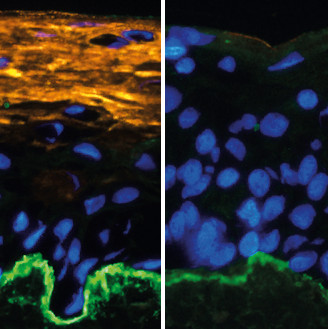
Markus Kretz | Long Non-Coding RNAs in Health and DiseaseLong non-coding RNAs (lncRNAs) have emerged as key regulators of gene expression in many different cellular pathways and have been implicated in numerous diseases, including several types of cancer. Using human organotypic epidermis as a model system, we are analyzing how lncRNAs interact with their protein- and RNA- binding partners as well as chromatin and thus help regulating the intricate balance between stem cells undergoing continual regeneration and highly differentiated cells forming the mature tissue environment. We are currently investigating how lncRNAs affect these cellular interactions in normal and diseased tissue and cells. |
 |
 |
Jan Medenbach | Post-Transcriptional Regulation of Gene ExpressionThe ability to sequence entire genomes and transcriptomes in ever shorter timeframes has far outpaced our understanding of how gene expression is controlled. Even though we have a firm knowledge of the general principles of gene expression, we are still far from a holistic and comprehensive understanding of the underlying regulation. It is one of the major challenges in modern biology to gain detailed understanding of the regulatory principles and networks that control gene expression. To better understand gene expression control, we are focusing on two major topics: 1. Gene regulation during cellular stress and its role in disease and resistance to therapy and 2. RNA-binding proteins in Drosophila development and sexual differentiation |
 |