ERC grants in the RCB
October 2023: ERC Synergy grant for Christine ZieglerSeptember 2023: ERC starting grant for Markus Jeschek
Publications
A publication list of the RCB members can be found hereSelected research highlights 2026
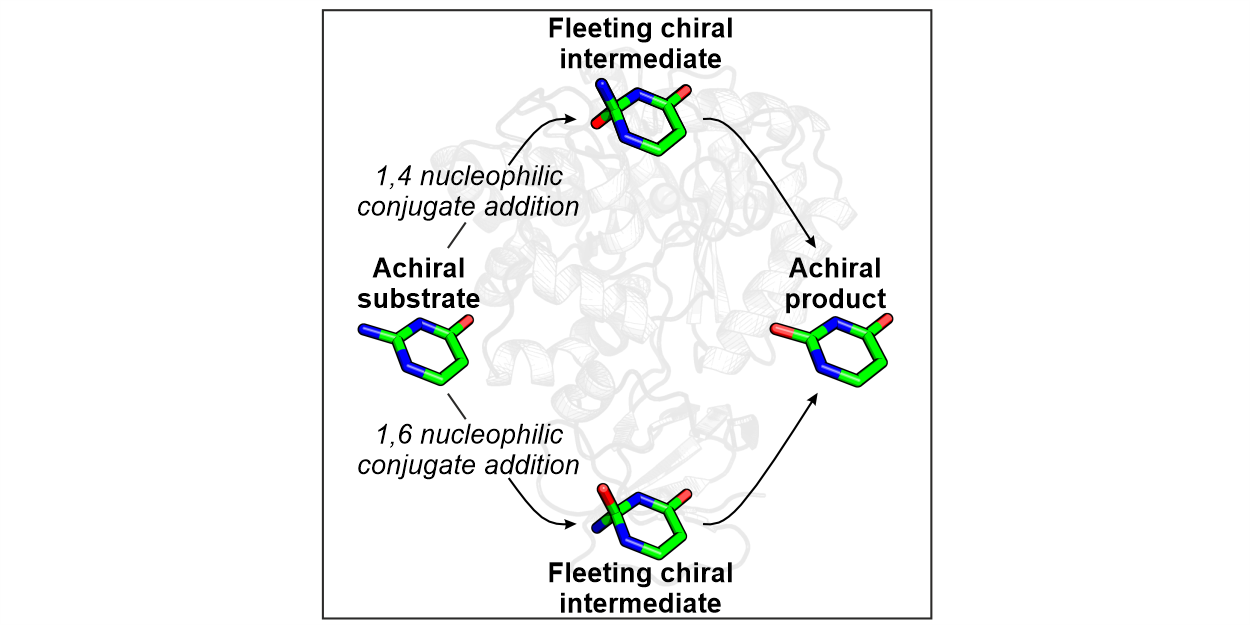 |
The sequencing of millions of open reading frames has identified numerous enzymes with unknown functions, which makes the deciphering of substrate specificity and promiscuity a central challenge in biochemistry. In this study, this problem has been addressed for enzymes from the amidohydrolase superfamily (AHS), specifically guanine deaminases (GuaD) and cytosine deaminases (CodA) and their homologues. We discovered that these enzymes utilize a reaction mechanism that proceeds either via 1,4 or 1,6 nucleophilic conjugate addition dictated by a specific glutamine residue located at different positions within the active site. This glutamine determines the orientation of a substrate and stabilizes Fleeting Chiral Intermediates (FCIs), transient chiral species that are generated in the course of the hydrolysis reaction although the AHS substrates and products are entirely achiral. Notably, the GuaD and CodA enzymes exhibit inverted enantioselectivity for these FCIs. The study also highlights that catalysis within these enzymes focuses on conserved core structures of a substrate rather than the entire molecule. This focus increases enzymatic promiscuity and evolvability, allowing these enzymes to rapidly adapt to xenobiotic compounds they have never previously encountered. The study was supported by a grant of the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
Reference:Drexler L, Fürtges TF, Rudack T, Sterner R (2026) On the Origin of Substrate Specificity of Enzymes from the Amidohydrolase Superfamily. Angew Chem Int Ed Engl. 65:e17873. doi:10.1002/anie.202517873 |
Selected research highlights 2025
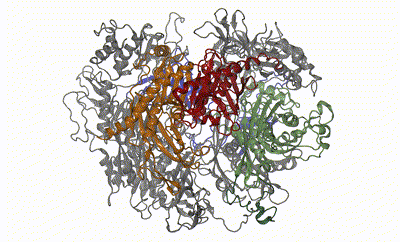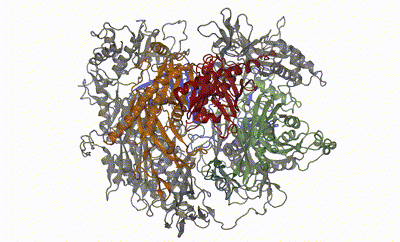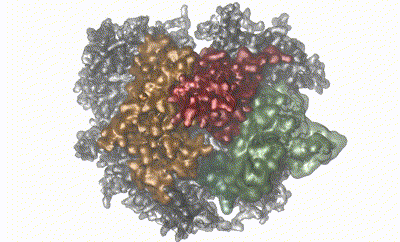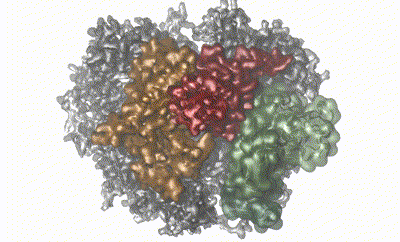 |
Large protein complexes are crucial in all aspects of biology. While detailed insights into the structure of such complexes can be obtained by, e.g., cryo-EM and computational methods, other properties, such as dynamics and weak molecular interactions, remain elusive and challenging to study. The eukaryotic RNA exosome is a fully asymmetric, decameric 410 kDa protein complex involved in 3'–5' RNA degradation. Using a combination of methyl- and 19F-labeling schemes and we employ nuclear magnetic resonance to demonstrate that RNA substrate is threaded through the central pore formed by the complex. We then characterize the millisecond timescale dynamics of an extended loop region that is invisible in our cryo-EM structure by NMR experiments. Both binding of the catalytic subunit and substrate interactions modify the dynamics of this loop. We then show that the loop region is in dynamic exchange between a rigid ‘closed’ state that is inconsistent with RNA binding and an ensemble of highly flexible ‘open’ states. Finally, employing activity assays we demonstrate that functionally the loop acts as a flexible plug that prevents RNA from accessing the catalytic site via an aberrant pathway. We thus demonstrate that NMR can be employed to obtain insights into interactions, conformations, and dynamics of large, asymmetric protein complexes. A combination of state-of-the-art NMR methods, structural methods, and MD simulations thus allows to go beyond purely static images of large, asymmetric protein complexes.
Reference:iJobst Liebau, Daniela Lazzaretti, Torben Fürtges, Anna Bichler, Michael Pilsl , Till Rudack, Remco Sprangers (2025) 4D structural biology-quantitative dynamics in the eukaryotic RNA exosome complex natr.commun. Aug 24;16(1):7896. doi:10.1038/s41467-025-62982-6 |
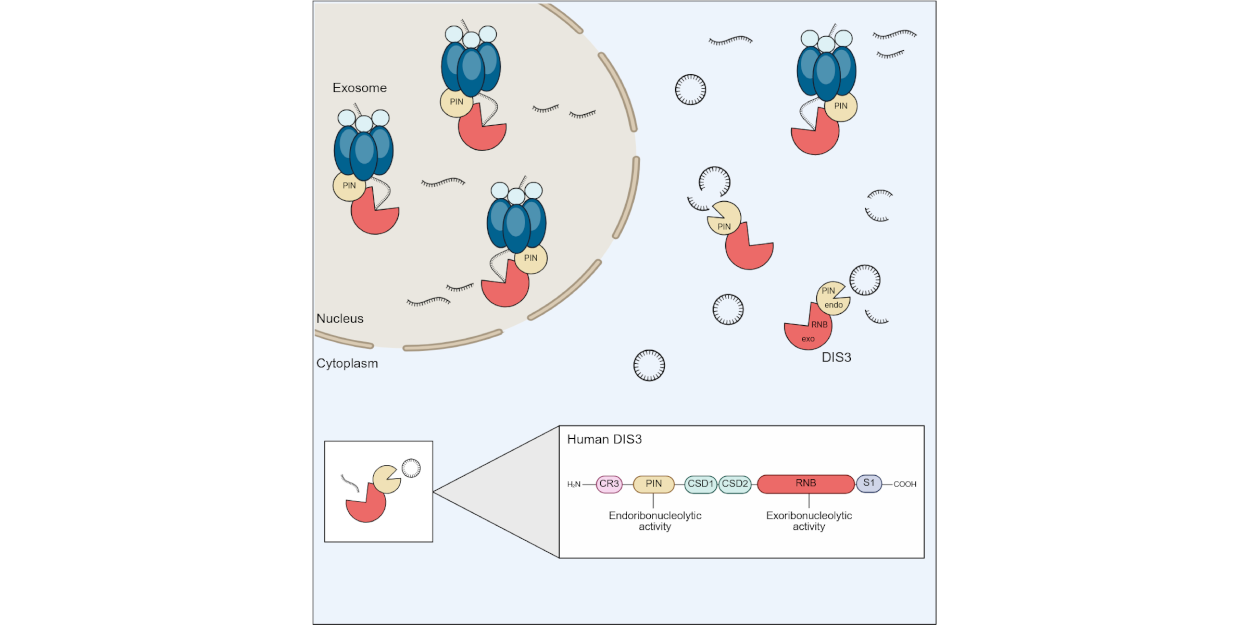 |
The ribonuclease DIS3 interacts through its PIN domain with the nuclear exosome and degrades linear RNA substrates using its exoribonuclease domain. However, the PIN domain is also an active endoribonuclease but cellular substrates are largely unknown. Here, we use a biochemical strategy to find ribonucleases that could degrade circular (circ)RNAs. Due to the lack of accessible ends, circRNAs are resistant to exonucleolytic cleavage and thus are more stable than linear RNAs. Using biochemical assays, we identify DIS3 as a candidate for circRNA degradation and demonstrate that it partially resides in the cytoplasm, where circRNAs are degraded. DIS3 shows cleavage activity towards a number of circRNAs and functions independently of the exosome core in vitro. Upon knock down of DIS3 in cell lines, selected circRNAs are moderately stabilized. We thus propose that cytoplasmic DIS3 functions as stand-alone enzyme independently of the exosome core and may contribute to circRNA turnover.
Reference:Latini C, Eichlinger J, Fuchs AL, Zhai SN, Ho-Xuan H, Lehmann G, Glažar P, Rajewsky N, Bruckmann A, Yang L, Sprangers R, Meister G (2025) Cytoplasmic DIS3 is an exosome-independent endoribonuclease with 0catalytic activity toward circular RNAs. Cell Rep. May 28;44(6):115769. doi:10.1016/j.celrep.2025.115769 |
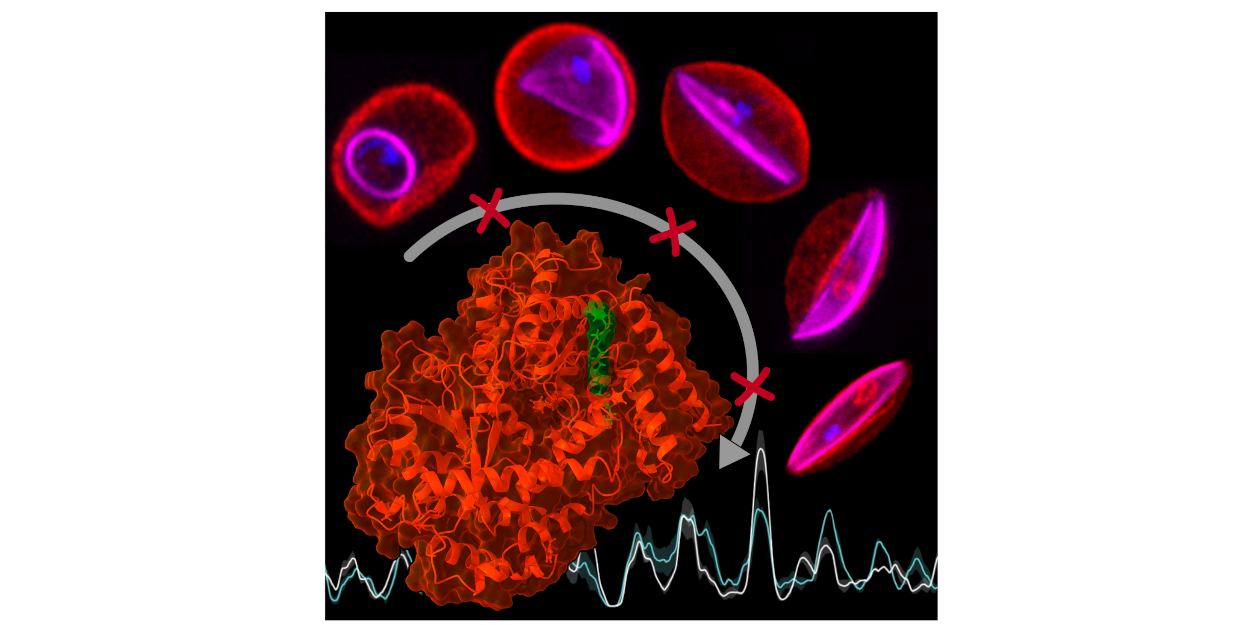 |
Malaria is caused by parasites of the genus Plasmodium, which is transmitted to humans through the bite of infected mosquitoes. Plasmodium falciparum, the deadliest of the malaria species, has a highly complex life cycle controlled by precise gene regulation. Understanding these regulatory processes is crucial to specifically combat the pathogen at different stages of development. Our new study reveals how regulatory proteins modulate genome-wide epigenetic mechanisms to control the precise timing of gene expression in the parasite. We show that the chromatin remodeler PfSnf2L is an essential regulator of “just-in-time” regulation of stage specifically expressed genes. The unique sequence and functional properties of PfSnf2L led to the identification of a highly specific inhibitor that only kills Plasmodium falciparum. A multidisciplinary approach, involving an innovative and unique screening platform for PfSnf2L inhibitors, genetic manipulation of malaria parasites and cutting-edge “OMICs” was used to uncover the role of PfSnf2L. This inhibitor represents a new class of antimalarials, potentially targeting all life cycle stages. The new findings not only advance basic research but could also offer practical applications. Malaria remains one of the greatest global health threats. In 2022, there were an estimated 247 million infections and over 600,000 deaths, mostly in sub-Saharan Africa. Innovative solutions such as targeting developmental regulation are therefore urgently needed to offer new treatment options. The study was carried out by an interdisciplinary, multinational team led by Prof. Gernot Längst (University of Regensburg, Germany) and Prof. Markus Meissner (Ludwig-Maximilians University, Munich, Germany) involving researchers from The University of Zürich (Switzerland), The Pennsylvania State University (USA) and The University of Glasgow (UK). It was supported by the DFG, German Research Foundation.
Reference:Watzlowik MT, Silberhorn E, Das S, Singhal R, Venugopal K, Holzinger S, Stokes B, Schadt E, Sollelis L, Bonnell VA, Gow M, Klingl A, Marti M, Llinás M, Meissner M, Längst G.. (2025) Plasmodium blood stage development requires the chromatin remodeller Snf2L. Nature Mar;639(8056):1069-1075. doi: 10.1038/s41586-025-08595-x |
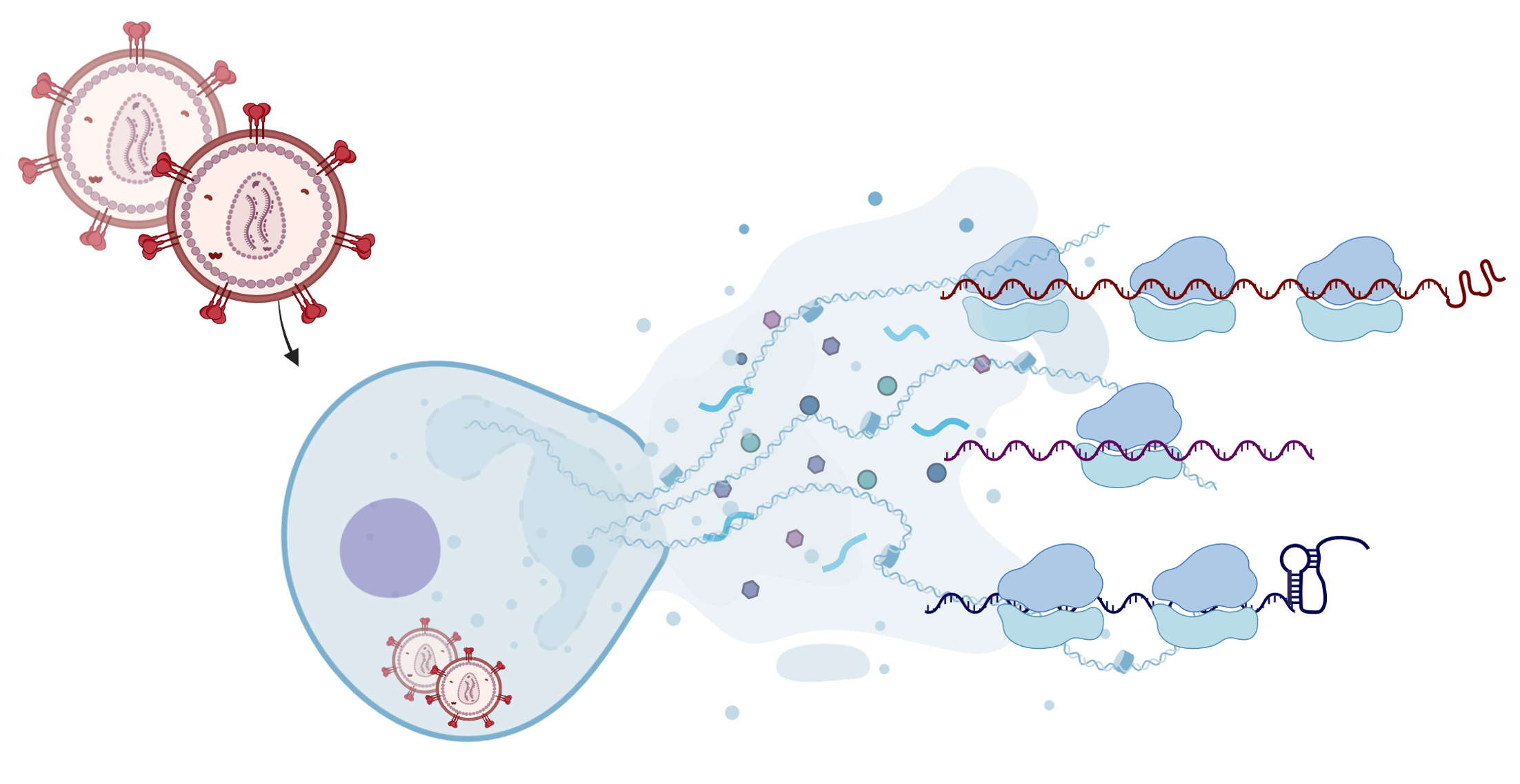 |
A recent study unveiled insights into how HIV-1, the virus responsible for AIDS, skillfully hijacks cellular machinery for its own survival. By dissecting the molecular interplay between the virus and its host, the researchers identified novel strategies that HIV-1 employs to ensure its replication while suppressing the host’s cellular defenses. The study was published in the journal Nature Structural and Molecular Biology. HIV-1, like other viruses, lacks the machinery to produce its own proteins and must rely on the host cell to translate its genetic instructions. After entering host cells, it seizes control of the translation process, which converts messenger ribonucleic acid (mRNA) into proteins. “In this study, we combined ribosome profiling, RNA sequencing and RNA structural probing to map the viral and host translational landscape and pausing during replication of the virus in unprecedented detail,” says corresponding author Neva Caliskan, who is heading the Department of Biochemistry III at the University of Regensburg.
Reference:Kibe A, Buck S, Gribling-Burrer AS, Gilmer O, Bohn P, Koch T, Mireisz CN, Schlosser A, Erhard F, Smyth RP, Caliskan N. (2025) The translational landscape of HIV-1 infected cells reveals key gene regulatory principles. Nat Struct Mol Biol." xxx-xx5 doi:10.1038/s41594-024-01468-3 |
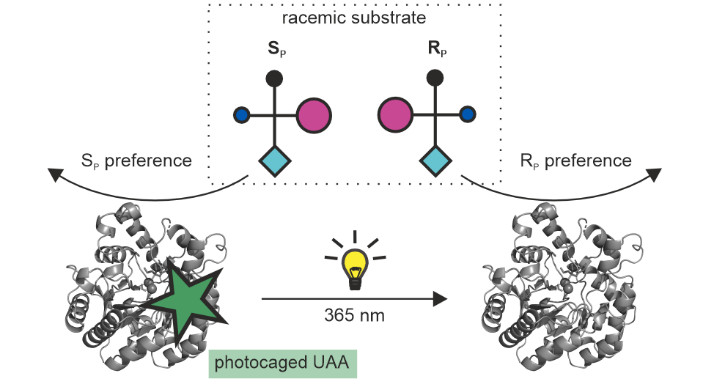 |
The external control of catalytic activity and substrate specificity of enzymes by light has aroused great interest in the fields of biocatalysis and pharmacology. Going beyond, we have photo-controlled enzyme stereoselectivity on the example of phosphotriesterase (PTE), which hydrolyzes a wide variety of racemic organophosphorus substrates. For this purpose, the photocaged unnatural amino acid o-nitrobenzyl-L-tyrosine (ONBY) was incorporated by genetic code expansion at the large subsite of the active center, together with additional mutations at the small subsite. Comparison of the enantioselectivities before and after decaging of ONBY using irradiation revealed the desired photo-induced inversion of enantioselectivity for several variants and substrates. A computational analysis showed that a complicated interplay between steric constraints and specific enzyme-substrate interactions is responsible for the observed effects. Our findings significantly broaden the scope of possibilities for the spatio-temporal control of enantioselective transformations using light in biocatalytic systems.
Reference:Hiefinger C, Zinner G, Fürtges TF, Narindoshvili T, Schindler S, Bruckmann A, Rudack T, Raushel FM, Sterner R. (2025) Photocontrolling the Enantioselectivity of a Phosphotriesterase via Incorporation of a Light-Responsive Unnatural Amino Acid. JACS AU Feb 5;5(2):858-870 doi:10.1021/jacsau.4c01106 |
Selected research highlights 2024
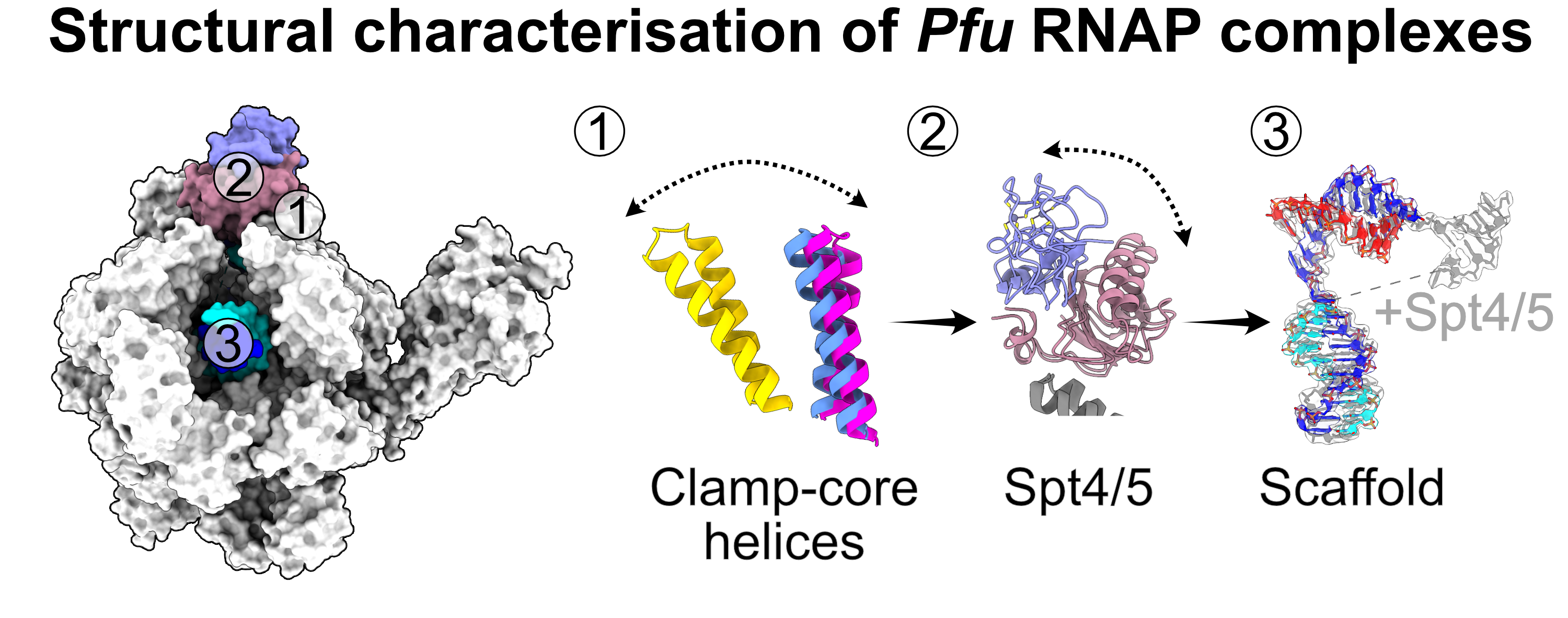 |
This study delves into the structural organization of archaeal transcription complexes. In a collaboration between the Microbiology and Structural Biochemistry group, we utilized single-particle cryo-electron microscopy, supplemented by single-molecule FRET and biochemcial analysis, to unravel the structure and dynamics of the RNA polymerase complexes from the archaeal model organism Pyrococcus furiosus. Our investigation sheds light on the influence of the transcription elongation factor Spt4/5 on the overall architecture and function of the RNA polymerase. This research unveils the broader role of Spt4/5 in transcription, potentially extending beyond its known function in elongation support
Reference:Tarău D, Grünberger F, Pilsl M, Reichelt R, Heiß F, König S, Urlaub H, Hausner W, Engel C, Grohmann D. (2024) Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment. Nucleic Acids Res 52(xi104):6017-6035 doi:10.1093/nar/gkae282 |
Selected research highlights 2023
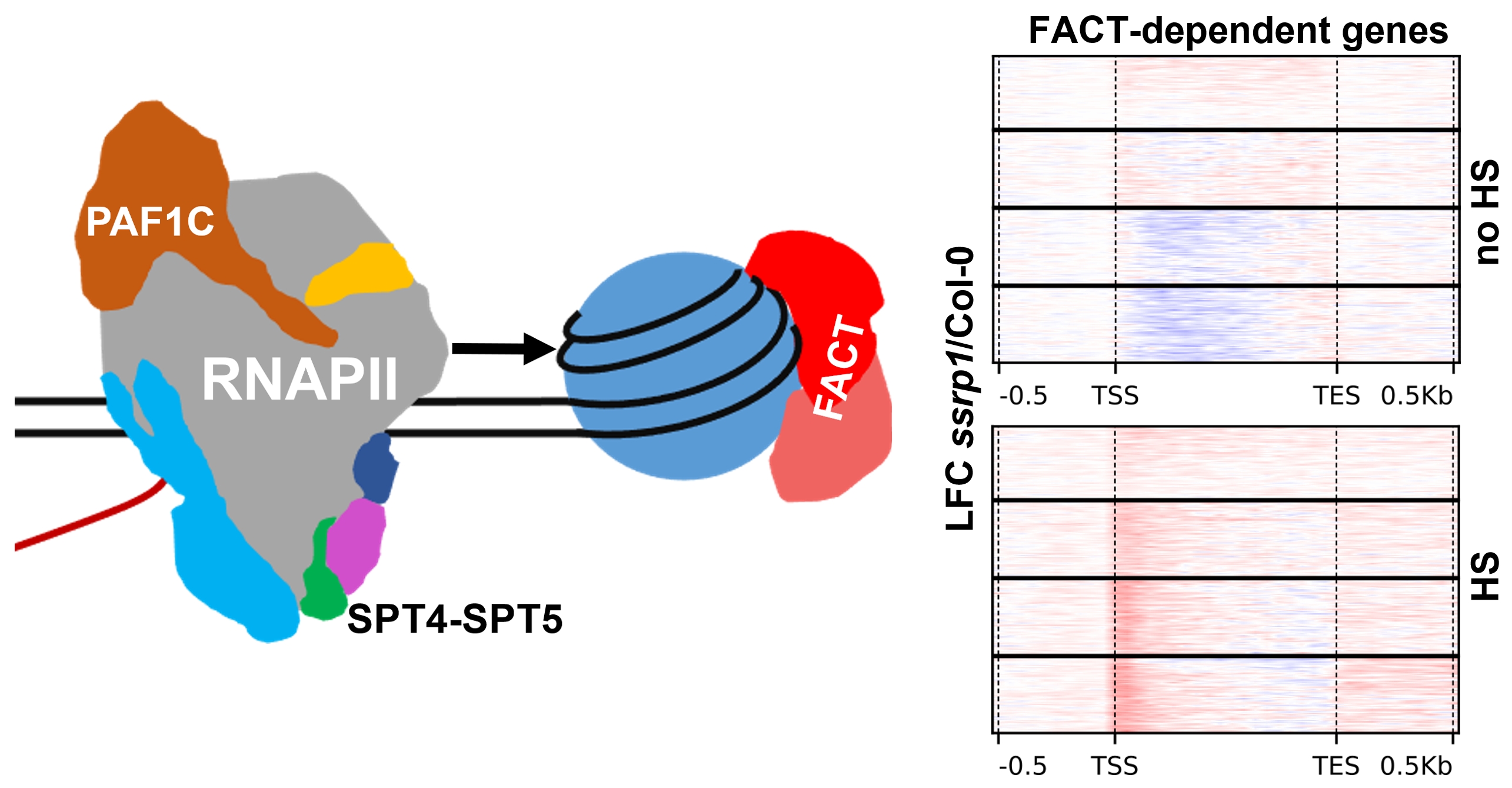 |
After transcriptional initiation by RNA polymerase II (RNAPII), the efficiency of mRNA synthesis is regulated by a variety of elongation factors (EFs). Mapping the genomic distribution of three EFs in Arabidopsis demonstrated that the occupancy of SPT4-SPT5 along transcribed regions closely resembles that of RNAPII-S2P, while the occupancy of FACT and PAF1C correlates with RNAPII-S5P. Under transcriptionally challenging heat stress (HS) conditions, PAF1C is required for transcription across intron/exon borders, whereas FACT is particularly required for transcriptional upregulation and to promote RNAPII transcription through +1 nucleosomes. Thus, the EFs are distinctly required during transcript elongation, particularly to accomplish transcriptional reprogramming.
Reference:Obermeyer S, Schrettenbrunner L, Stöckl R, Schwartz U, Grasser KD. (2023) Different elongation factors distinctly modulate RNA polymerase II transcription in Arabidopsis Nucleic Acids Res 51(21):11518-11533 doi:10.1093/nar/gkad825 |
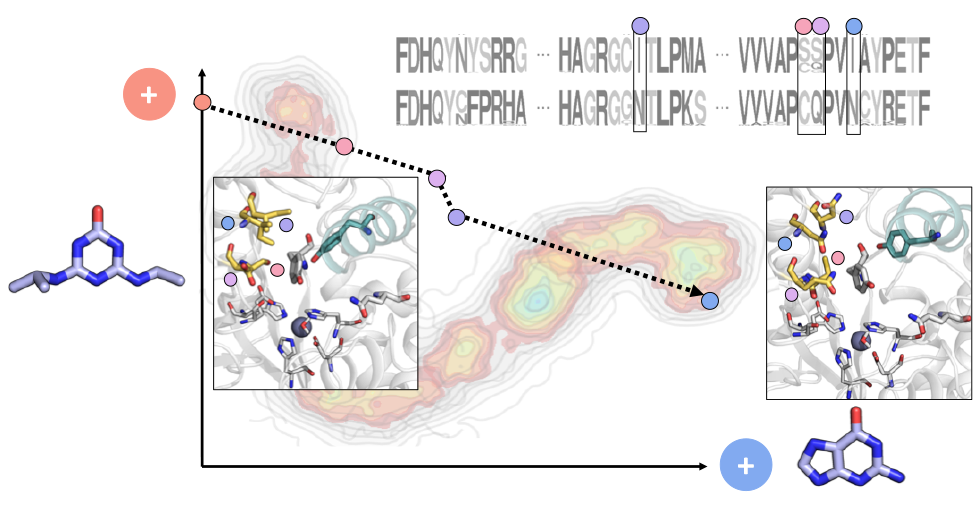 |
It has been generally assumed that it took nature billions of years of evolution to create highly efficient and specific enzymes. However, the discovery of metabolic pathways that allow for the rapid degradation of modern anthropogenic compounds has questioned this view and suggested that novel enzymes can evolve from ancestral ones within a very short time. Here, such fast molecular evolution has been experimentally retraced by protein engineering. Specifically, the hydroxyatrazine ethylaminohydrolase, which is a “young” enzyme involved in the decomposition of the herbicide atrazine, and its putative guanine deaminase precursor, which is an “old” enzyme from primary metabolism, could be interconverted into each other by only four amino acid exchanges.
Reference:Busch M, Drexler L, Mahato DR, Hiefinger C, Osuna S, Sterner R (2023) Retracing the Rapid Evolution of an Herbicide-Degrading Enzyme by Protein Engineering ACS Catalysis 13:15558-15571 doi:10.1021/acscatal.3c04010 |
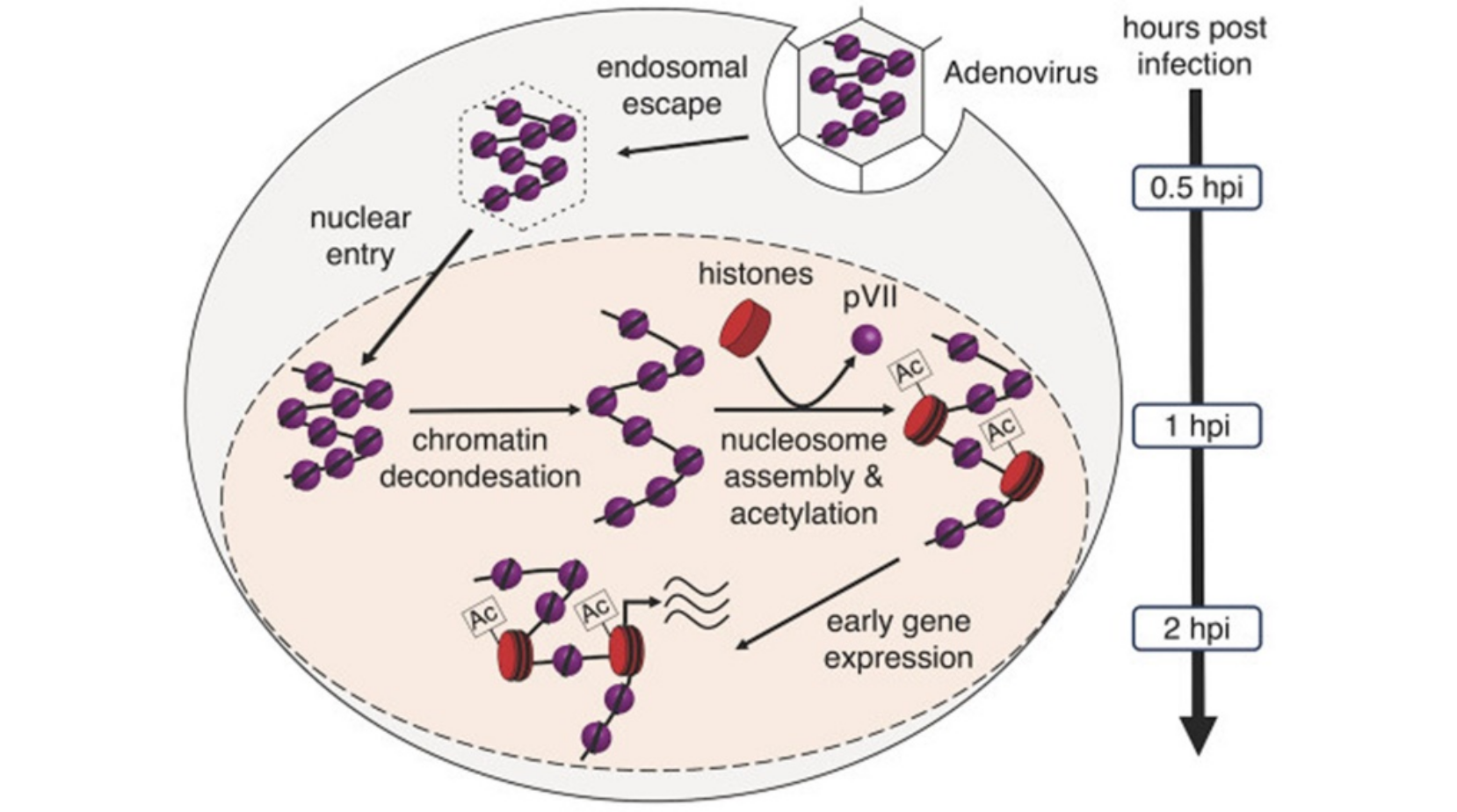 |
Adenoviruses, known for their low pathogenicity and technological approachability, have become instrumental in many therapeutic applications, including as vaccination vector platform during the recent SARS-CoV-2 pandemic. Central to their efficacy is the remarkably efficient delivery and expression of their genetic material. Two scientific teams led by Gernot Laengst from the University of Regensburg and Harald Wodrich from the University of Bordeaux have obtained insight into this process in unprecedented detail. Employing advanced microscopy techniques and genome structure analyses, they traced the early stages of adenoviral infection in human cells. This approach allowed a detailed description of the viral genome intricately wrapped with the viral pVII protein within the incoming virion. Furthermore, their investigation revealed the transformation of the viral chromatin structure, converting the densely packed genome into an exceptionally efficient template for gene expression as it traverses from the cell surface to the cell nucleus.
Reference:Schwartz U, Komatsu T, Huber C, Lagadec F, Baumgartl C, Silberhorn E, Nuetzel M, Rayne F, Basyuk E, Bertrand E, Rehli M, Wodrich H, Laengst G. (2023) Changes in adenoviral chromatin organization precede early gene activation upon infection EMBO J. e114162 doi:10.15252/embj.2023114162 |
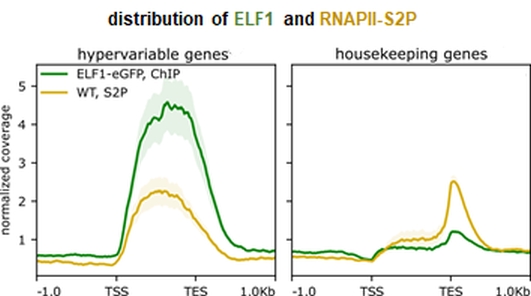 |
Elongation factors modulate the efficiency of mRNA synthesis by RNA polymerase II (RNAPII) in the context of chromatin, contributing to implement proper gene expression programs. The Arabidopsis zinc-finger protein ELF1 interacts with DNA and histones in vitro and with the RNAPII elongation complex in vivo. Analysis of a CRISPR-Cas9 mediated gene editing mutant of ELF1 revealed distinct genetic interactions with mutants deficient in other elongation factors. Moreover, ELF1 associated with genomic regions actively transcribed by RNAPII, and ELF1 occupied only ~33% of the RNAPII transcribed loci with preference for inducible rather than constitutively expressed genes. Thus, Arabidopsis ELF1 shares several characteristic attributes with RNAPII transcript elongation factors.
Reference:Markusch H, Michl-Holzinger P, Obermeyer S, Thorbecke C, Bruckmann A, Babl S, Längst G, Osakabe A, Berger F, Grasser KD. (2023) Elongation factor 1 is a component of the Arabidopsis RNA polymerase II elongation complex and associates with a subset of transcribed genes New Phytol 238(1): 113-124 doi:10.1111/nph.18724 |
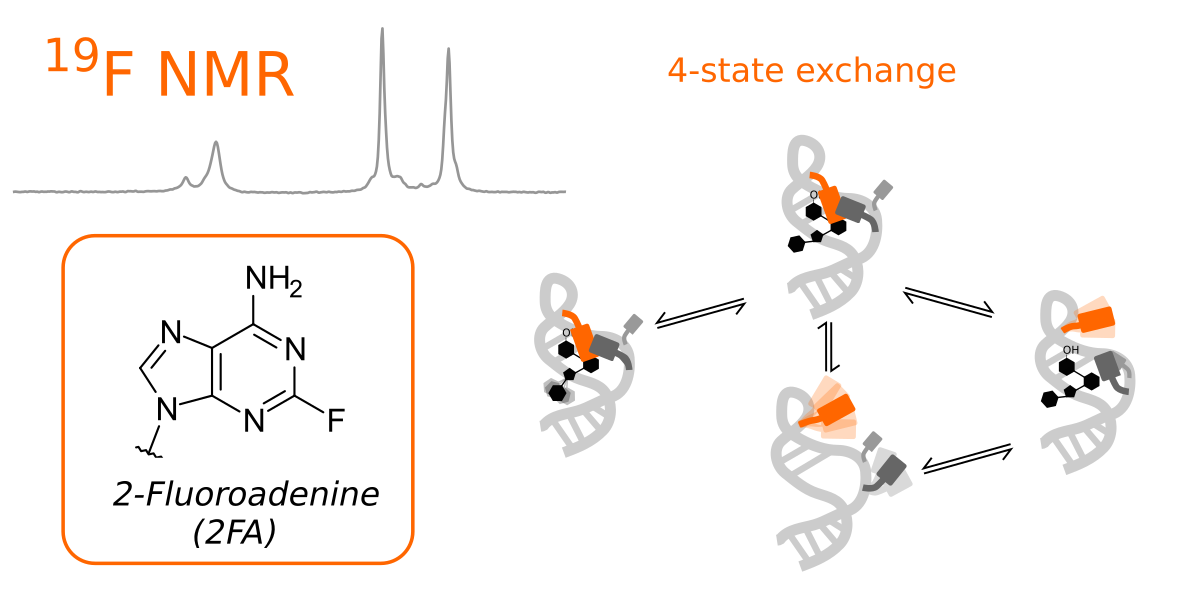 |
Molecular motions are tightly coupled with function. Here we use a unique combination of five different fluorine NMR relaxation methods to reveal the dynamics in three structurally similar riboswitch:ligand complexes. Our data displays unexpectedly complex exchange topologies with up to four structurally different states. Importantly, the associated kinetic and thermodynamics parameters could be accurately determined.
Reference:Overbeck JH, Vögele J, Nussbaumer F, Duchardt-Ferner E, Kreutz C, Wöhnert J, Sprangers R. (2023) Multi-Site Conformational Exchange in the Synthetic Neomycin-Sensing Riboswitch Studied by 19 F NMR Angew Chem Int Ed e202218064 doi:10.1002/anie.202218064 |
Selected research highlights 2022
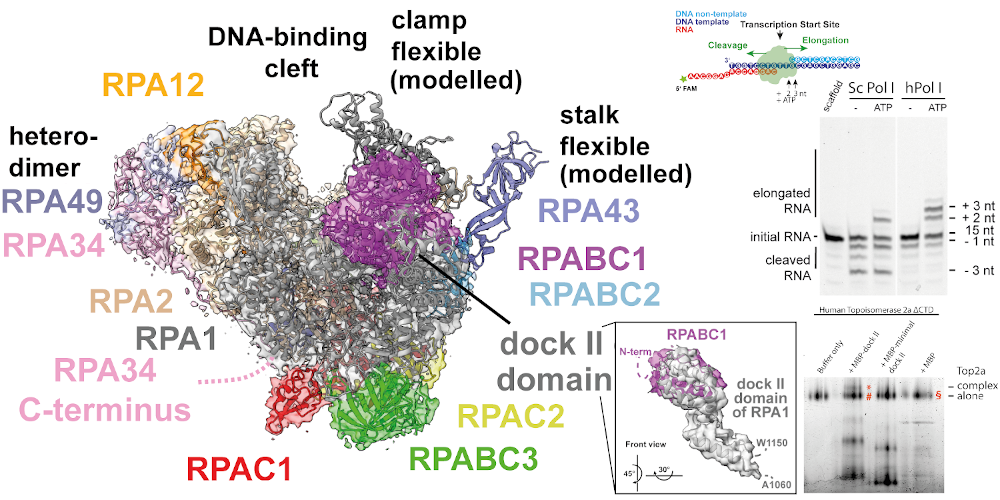 |
Transcription of the ribosomal RNA (rRNA) precursor by RNA polymerase (Pol) I is a major determinant of cellular growth, and dysregulation is observed in many cancer types. We generated a human cell line allowing for the purification of human Pol I for its structural and functional characterization. In contrast to its yeast homologue, human Pol I contains a single-subunit stalk, shows a reduced proofreading activity and carries an additional domain within its largest subunit. This “dock II” domain resembles a truncated HMG box and may serve as a transcription factor binding platform. We find evidence that Topoisomerase 2a could be recruited to Pol I via this domain. Despite overall conservation from yeast to human, specific features suggest an accumulating specialization of Pol I defining its adaptation to the unique task of rRNA synthesis throughout evolution.
Reference:Daiß JL, Pilsl M, Straub K, Bleckmann A, Höcherl M, Heiss FB, Abascal-Palacios G, Ramsay EP, Tlučková K, Mars J-C, Fürtges T, Bruckmann A, Rudack T, Bernecky C, Lamour V, Panov K, Vannini A, Moss T, Engel C. (2022) The human RNA polymerase I structure reveals an HMG-like docking domain specific to metazoans Life Science Alliance 5(11) doi:10.26508/lsa.202201568 |
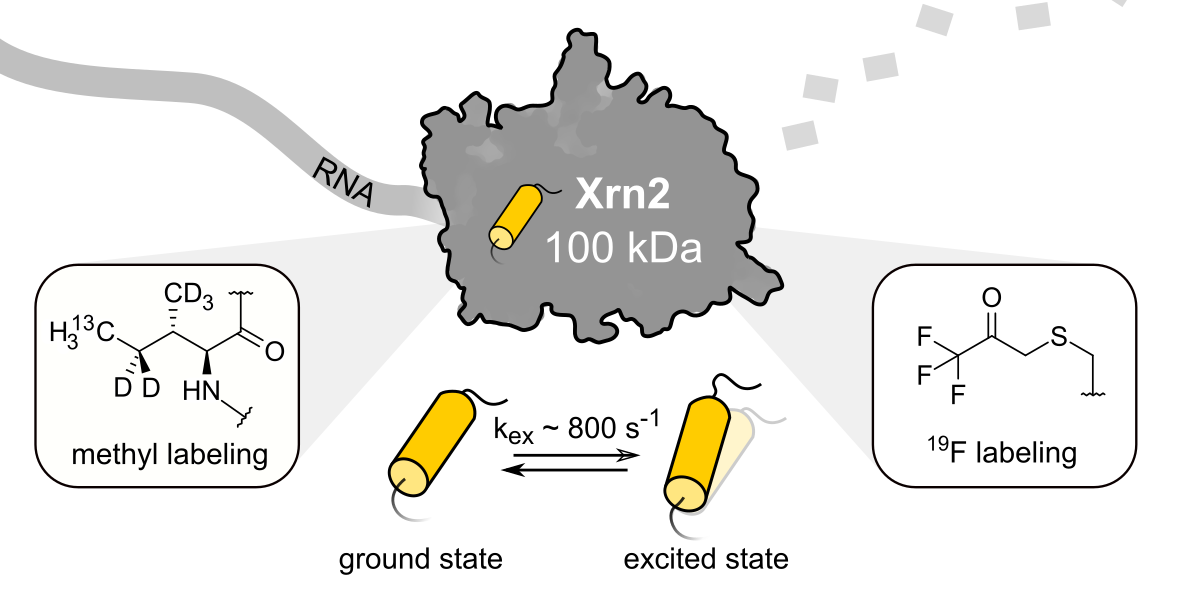 |
The essential eukaryotic 5′→3′ exoribonuclease Xrn2 contributes to cellular RNA homeostasis and is involved in RNA degradation and processing as well as transcription termination. Here, we use state-of-the-art nuclear magnetic resonance (NMR) methods probing methyl groups and trifluoromethyl-containing moieties to quantitatively study the dynamics of a wild type 100 kDa core construct of Xrn2 from the thermophilic eukaryote Chaetomium thermophilum. We show that the apo enzyme is highly dynamic in the vicinity of the catalytic center, where the excited state resembles a pre-hydrolysis state that is stabilized upon ligand binding. Our results corroborate and quantify a model derived from static X-ray and cryo-EM structures, in which extensive motions accompany the catalytic cycle of Xrn2.
Reference:Overbeck JH, Stelzig D, Fuchs AL, Wurm JP, Sprangers R. (2022) Observation of conformational changes that underlie the catalytic cycle of Xrn2. Nat Chem Biol doi:10.1038/s41589-022-01111-6 |
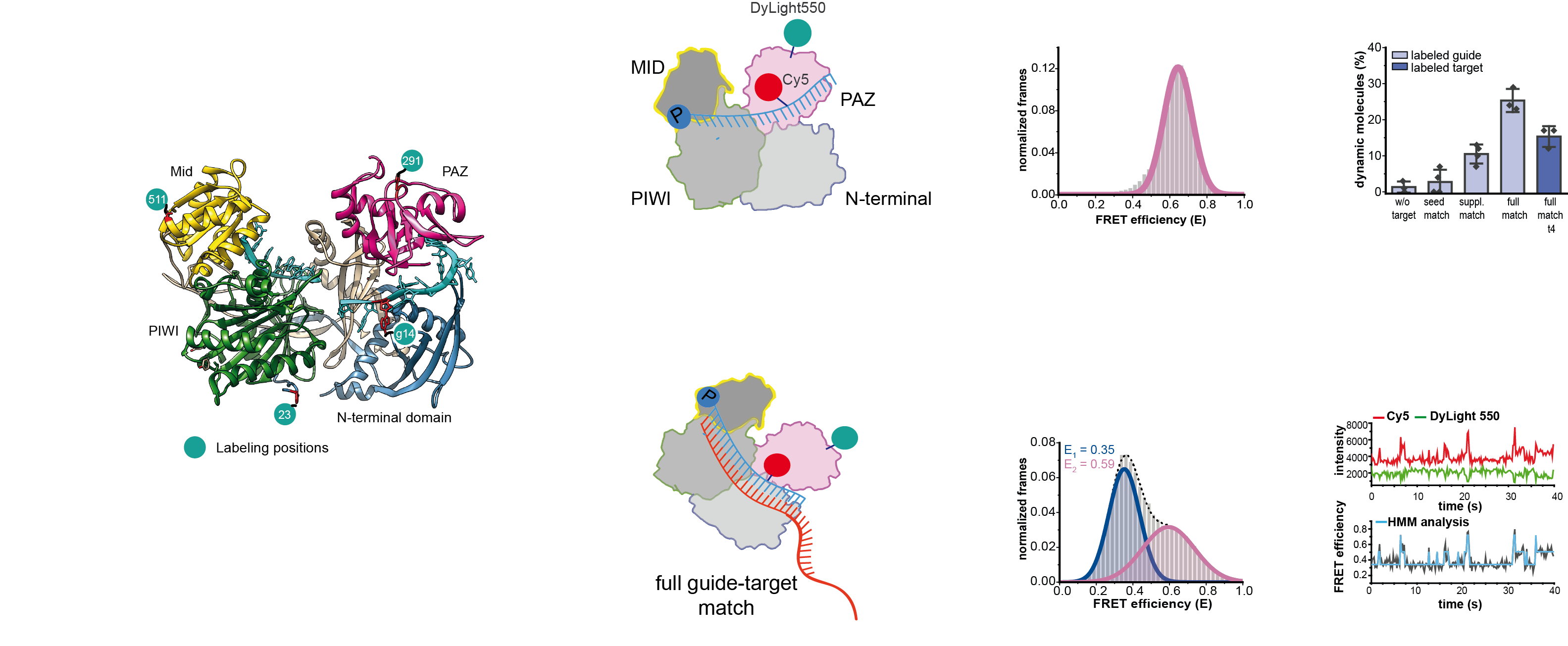 |
Argonaute proteins are the key players in RNAi. Human Argonaute 2 (hAgo2) with its catalytic slicing activity raised remarkable scientific and biotechnological interest with far-reaching implications for RNA-based therapy. In this publication, we succeeded to fluorescently label native hAgo2 site-specifically allowing for comprehensive intramolecular FRET measurements unlocking the full potential of smFRET studies to access conformational heterogeneity and transient states. We performed time-resolved single-molecule fluorescence resonance energy transfer (smFRET) measurements with apo hAgo2 and a wide range of defined hAgo2-guide and hAgo2-guide/target complexes in solution. We expand the structural and mechanistic framework of hAgo2 and hAgo2-RNA complexes showing among others (i) that the 3’-end of the guide can dynamically re-associate with the PAZ domain thereby preventing degradation of the guide, (ii) that Ago2-guide/target complex that resemble substrates for target-directed miRNA degradation (TDMD) adopt a unique conformation not detected so far using structural methods and (iii) the structural difference of the Ago-processed niRNA451 to canonical hAgo2-guide/target complexes.
Reference:Willkomm S, Jakob L, Kramm K, Graus V, Neumeier J, Meister G, Grohmann D. (2022) Single-molecule FRET uncovers hidden conformations and dynamics of human Argonaute 2 Nat Commun. 13(1):3825 doi: 10.1038/s41467-022-31480-4 |
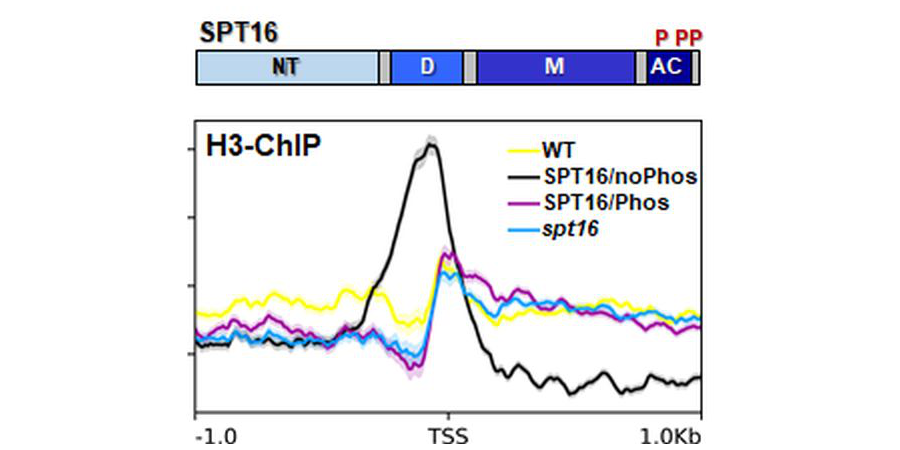 |
The histone chaperone FACT, consisting of SSRP1 and SPT16, contributes to dynamic nucleosome rearrangements. SPT16 was isolated from Arabidopsis cells and three phosphorylated Ser/Thr residues were identified in the acidic region by mass spectrometry. A non-phosphorylatable version of SPT16 displayed reduced histone binding and proved inactive in complementing the phenotypes of spt16 mutants. In plants expressing the non-phosphorylatable SPT16 version, at a subset of genes enrichment of histone H3 was detected, directly upstream of transcriptional start sites (TSSs). Thus, some genes require phosphorylation of SPT16 for establishing the correct nucleosome occupancy at the TSS of active genes.
Reference:Michl-Holzinger P, Obermeyer S, Markusch H, Pfab A, Ettner A, Bruckmann A, Babl S, Längst G, Schwartz U, Tvardovskiy A, Jensen ON, Osakabe A, Berger F, Grasser KD (2022) Phosphorylation of the FACT histone chaperone subunit SPT16 affects chromatin at RNA polymerase II transcriptional start sites in Arabidopsis. Nucleic Acids Res. 50(9): 5014-5028 doi:10.1093/nar/gkac293 |
Selected research highlights 2021
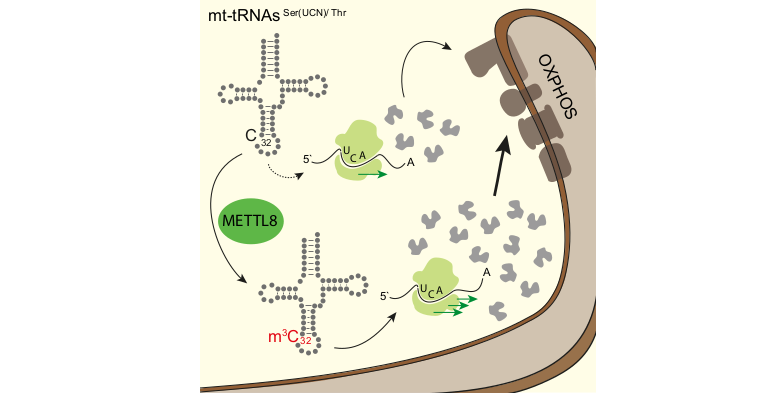 |
The m3C methyltransferase METTL8 methylates C32 on mitochondrial tRNAs mt-tRNASer(UCN) and mt-tRNAThr. This modification affects mitochondrial translation efficiency and respiratory chain activity. Schöller et al. observe stalled ribosomes on specific mt-tRNASer(UCN) codons in the absence of METTL8 leading to misbalancing of respiratory chain complexes potentially relevant for pancreatic cancer.
Reference:Schöller E, Marks J, Marchand V, Bruckmann A, Powell CA, Reichold M, Mutti CD, Dettmer K, Feederle R, Hüttelmaier S, Helm M, Oefner P, Minczuk M, Motorin Y, Hafner M, Meister G. (2021) Balancing of mitochondrial translation through METTL8-mediated m3C modification of mitochondrial tRNAs. Mol Cell. S1097-2765(21)00910-2 doi:10.1016/j.molcel.2021.10.018 |
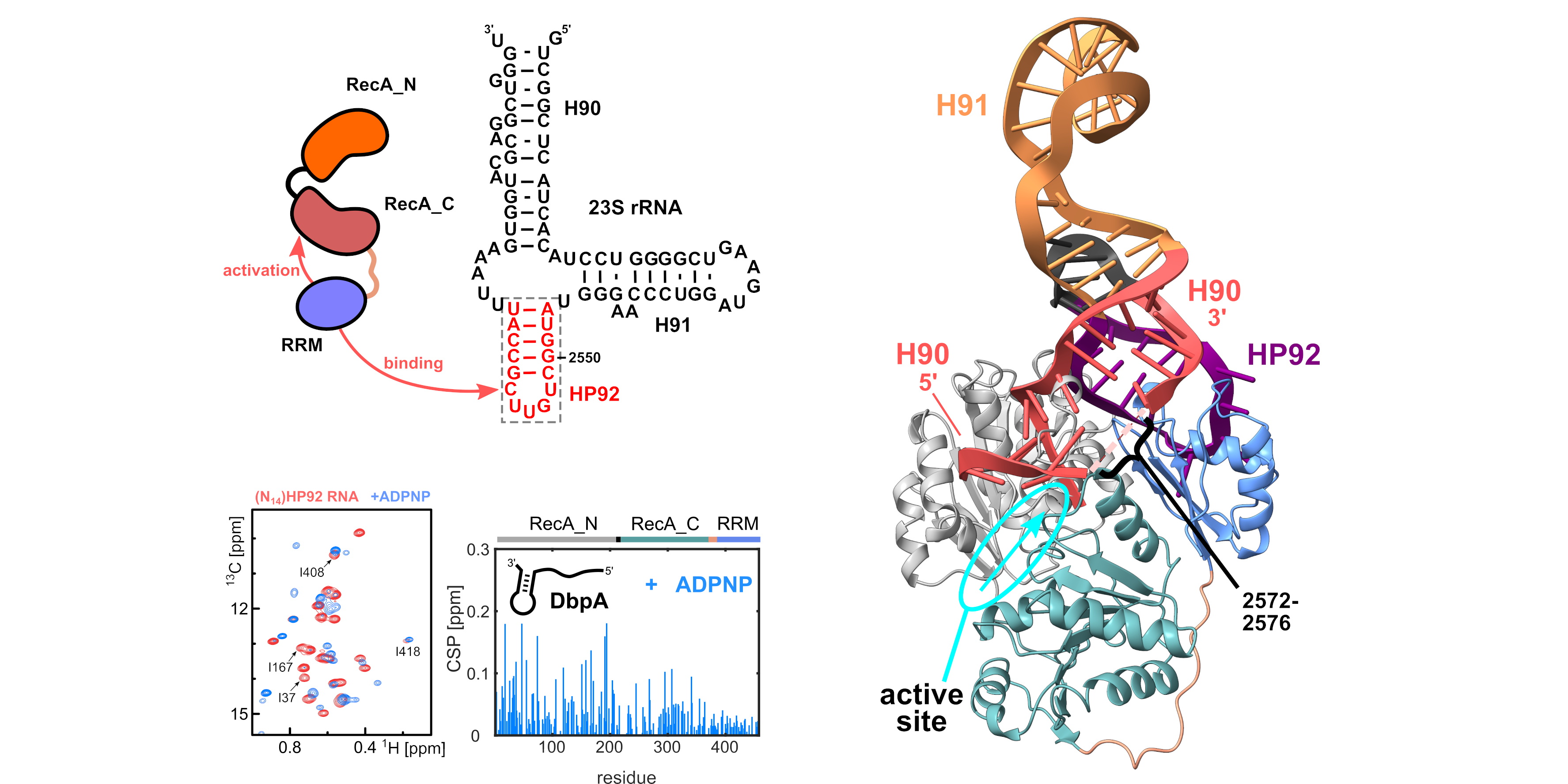 |
DEAD-box RNA helicases resolve misfolded RNA structures in an ATP-dependent manner and are involved in most aspects of RNA biology. In this study we elucidated the molecular mechanism behind the activation of the DEAD-box helicase DbpA and identified the RNA target region of DbpA during ribosome maturation. DbpA is activated upon binding to a specific region of the ribosomal RNA in maturing ribosomes. This selective activation of DbpA ensures that DbpA is only active on its RNA target sequence within the maturing ribosome and also prevents the wasteful hydrolysis of ATP in the absence of the maturing ribosome. NMR-based structural and interactions studies reveal that the activation critically depends on the precise positioning of the ribosomal RNA by an ancillary RNA binding domain of Dbpa, which leads to the thermodynamic stabilization of the catalytically active conformation of DbpA. Our results regarding the RNA target region also provide important insights into the function of DbpA during ribosome biogenesis.
Reference:Wurm JP, Glowacz KA, Sprangers R (2021) Structural basis for the activation of the DEAD-box RNA helicase DbpA by the nascent ribosome. Proc Natl Acad Sci USA 118(35):e2105961118 doi:10.1073/pnas.2105961118 |
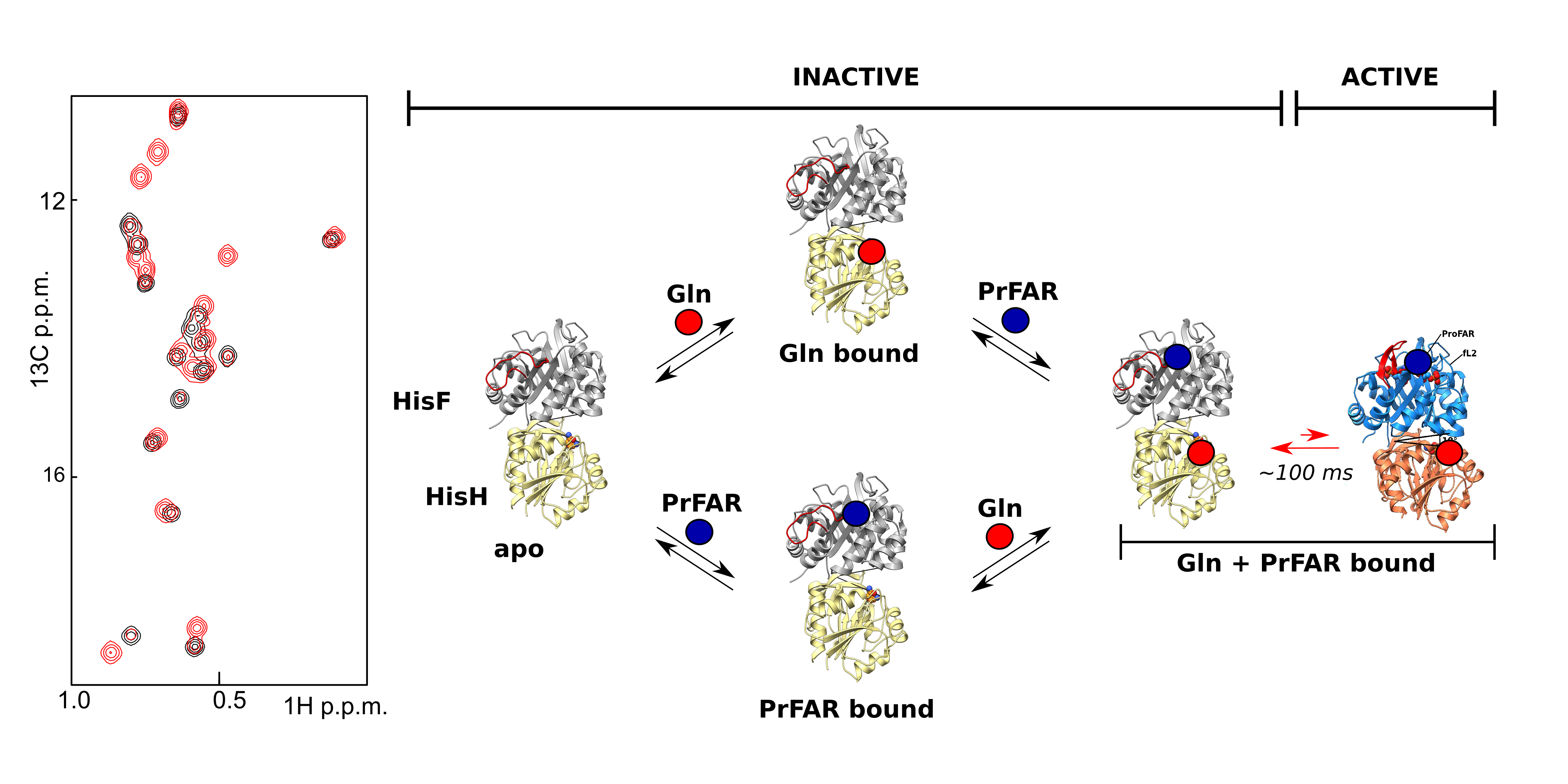 |
Allosteric regulation is a ubiquitous phenomenon in biochemistry, nevertheless, the underlying mechanisms are not well understood. Here, we elucidate the molecular basis of a long range, allosteric activation mechanism in the heterodimeric bienzyme complex imidazole glycerol phosphate synthase (HisFH). Using X-ray crystallography we determined the active conformation of the complex, where the oxyanion hole in the HisH active site is fully formed. This structure rationalizes the observed 4,500-fold allosteric activation compared to the inactive conformation. Based on methyl TROSY NMR experiments we show that the active and the inactive ground conformations are in a dynamic equilibrium in solution. Importantly, we show that the HisFH turnover rates directly correlate with the population of the active conformation which is in accordance with the ensemble model of allostery.
Reference:Wurm JP, Sung S, Kneuttinger AC, Hupfeld E, Sterner R, Wilmanns M, Sprangers R. (2021) Molecular basis for the allosteric activation mechanism of the heterodimeric imidazole glycerol phosphate synthase complex. Nat Commun. 12(1):2748 doi:10.1038/s41467-021-22968-6 |
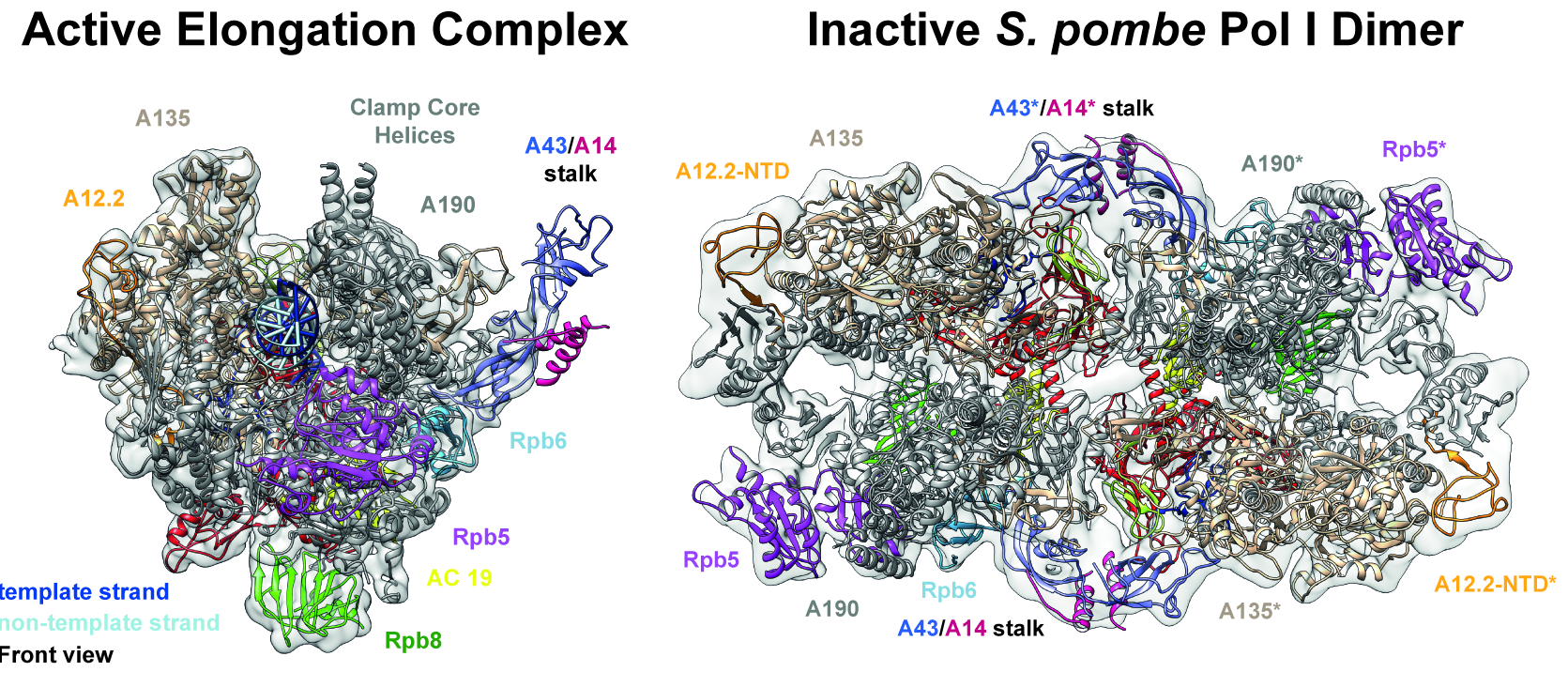 |
RNA polymerase (Pol) I transcribes the ribosomal RNA precursor in all eukaryotes. We used single particle cryo-EM to solve the structure of Pol I from fission yeast Schizosaccharomyces pombe in three different states. Our results demonstrate that regulatory mechanisms specific to Pol I are shared among organisms despite structural divergence. Consequently, ‘activation by contraction’ and ‘hibernation by dimerization’ of Pol I are observed in our reconstructions and confirmed by in vitro biophysical analysis. Our findings highlight the divergent nature of Pol I transcription systems from their Pol II and III counterparts and allow discussing evolutionary conserved features and mechanisms.
Reference:Heiss FB, Daiß JL, Becker P, Engel C. (2021) Conserved strategies of RNA polymerase I hibernation and activation. Nat Commun. 12(1):758 doi:10.1038/s41467-021-21031-8 |
 |
In this study, we have analysed the in vivo role of the almost universally conserved ribosomal RNA dimethyltransferase KsgA/Dim1 homolog in archaea. Combining structure-based functional studies across evolutionary divergent organisms, we provide evidence on how rRNA structure sequence variability (re-)shapes the KsgA/Dim1-dependent rRNA modification status. Collectively, our study provides additional understandings into principles of molecular functional adaptation, and further evolutionary and mechanistic insights into an almost universally conserved step of ribosome synthesis.
Reference:Knüppel R, Trahan C, Kern M, Wagner A, Grünberger F, Hausner W, Quax TEF, Albers SV, Oeffinger M, Ferreira-Cerca S. (2021) Insights into synthesis and function of KsgA/Dim1-dependent rRNA modifications in archaea. Nucleic Acids Res. 49(3):1662-1687 doi:10.1093/nar/gkaa1268 |
Selected research highlights 2020
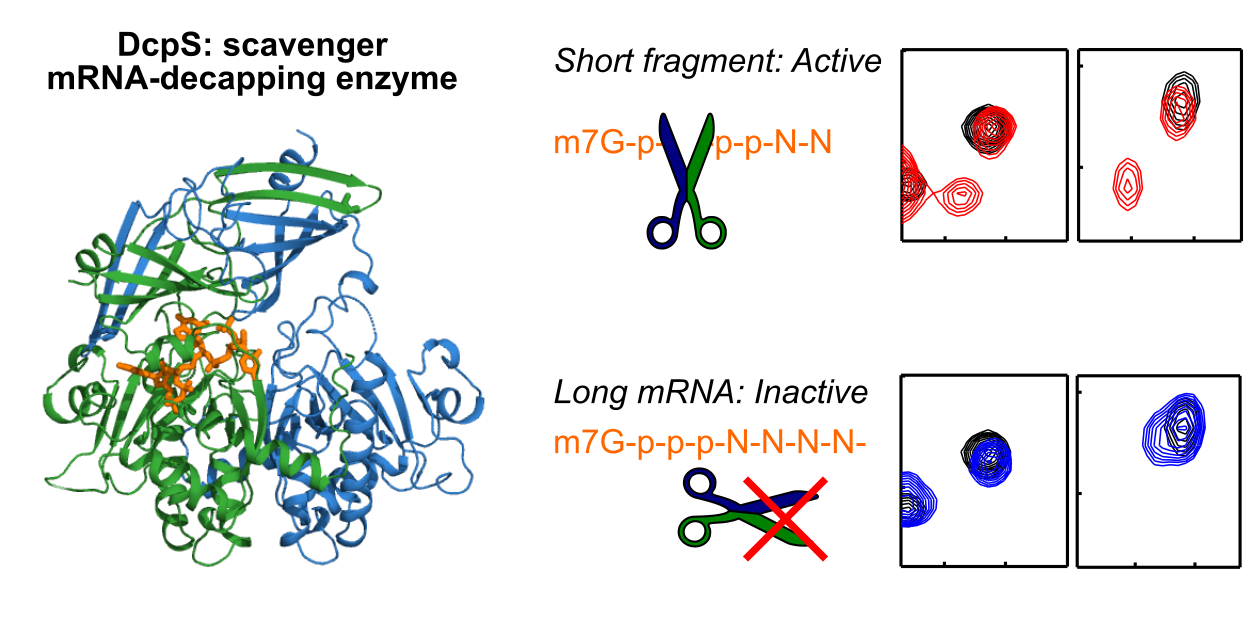 |
In eukoryotes, 3′ to 5′ mRNA degradation is a major pathway to reduce mRNA levels and, thus, an important means to regulate gene expression. Herein, messenger RNA (mRNA) is hydrolyzed from the 3′ end by the exosome complex, producing short capped RNA fragments, which are decapped by DcpS. Our data show that DcpS is only active on mRNA that have undergone prior processing by the exosome. This DcpS selection mechanism is conserved from yeast to humans and is caused by the inability of the enzyme to undergo structural changes that are required for the formation of a catalytically active state around long mRNA transcripts. Our work thus reveals the mechanistic basis that ensures an efficient interplay between DcpS and the exosome.
Reference:Fuchs AL, Wurm JP, Neu A, Sprangers R (2020) Molecular basis of the selective processing of short mRNA substrates by the DcpS mRNA decapping enzyme Proc Natl Acad Sci USA 117(32):19237-19244 doi:10.1073/pnas.2009362117 |
 |
Resistance of cancer cells against therapeutic agents is a major cause of treatment failure, especially in recurrent diseases. In a collaborative effort, we identified a novel mechanism of chemoresistance which has now been published in ‘Nature Communications’. It is driven by the Unfolded Protein Response (UPR), a cellular stress response pathway that alters gene expression and cellular metabolism to promote cell survival under stress.
Reference:Reich S, Nguyen CDL, Has C, Steltgens S, Soni H, Coman C, Freyberg M, Bichler A, Seifert N, Conrad D, Knobbe-Thomsen CB, Tews B, Toedt G, Ahrends R, Medenbach J. (2020) A multi-omics analysis reveals the unfolded protein response regulon and stress-induced resistance to folate-based antimetabolites. Nat Commun. 10;11(1):2936 doi:10.1038/s41467-020-16747-y |
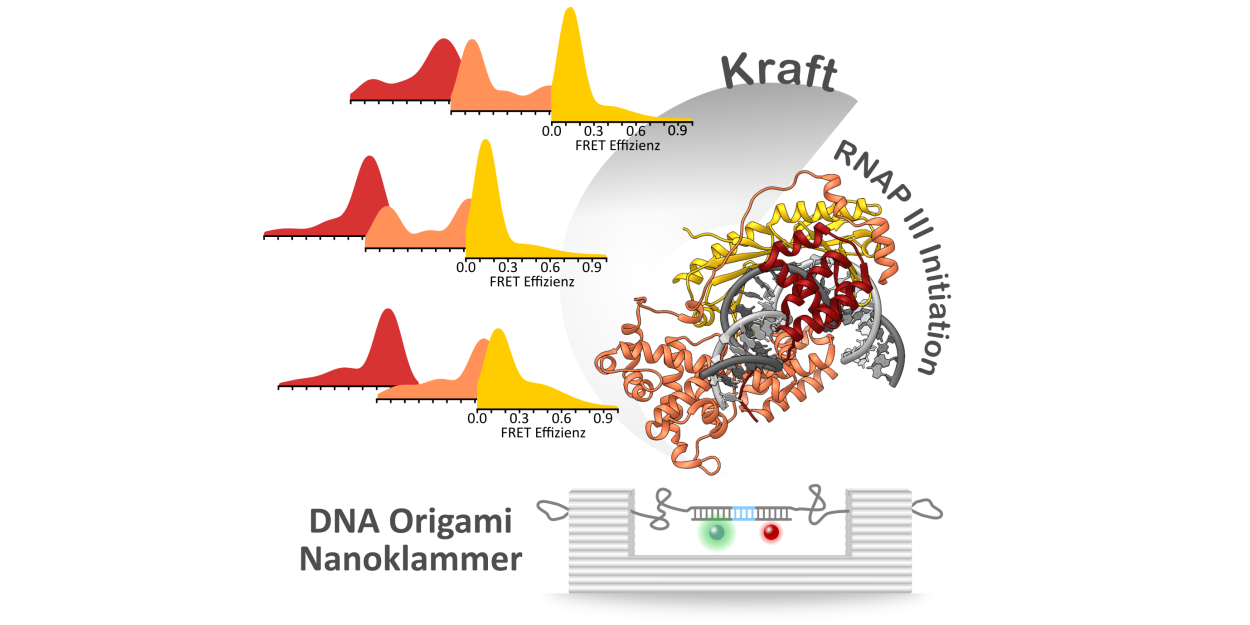 |
We made use of a DNA origami-based force clamp to follow the assembly of human initiation complexes in the RNAP II and RNAP III systems at the single-molecule level under piconewton forces. We demonstrate that TBP-DNA complexes are force-sensitive and TFIIB is sufficient to stabilise TBP on a strained promoter. In contrast, Bdp1 is the pivotal component that ensures stable anchoring of initiation factors, and thus the polymerase itself, in the RNAP III system. Thereby, we offer an explanation for the crucial role of Bdp1 for the high transcriptional output of RNAP III.
Reference:Kramm K, Schröder T, Gouge J, Vera AM, Gupta K, Heiss FB, Liedl T, Engel C, Berger I, Vannini A, Tinnefeld P, Grohmann D. (2020) DNA origami-based single-molecule force spectroscopy elucidates RNA Polymerase III pre-initiation complex stability. Nat Commun. 11(1):2828 doi:10.1038/s41467-020-16702-x |
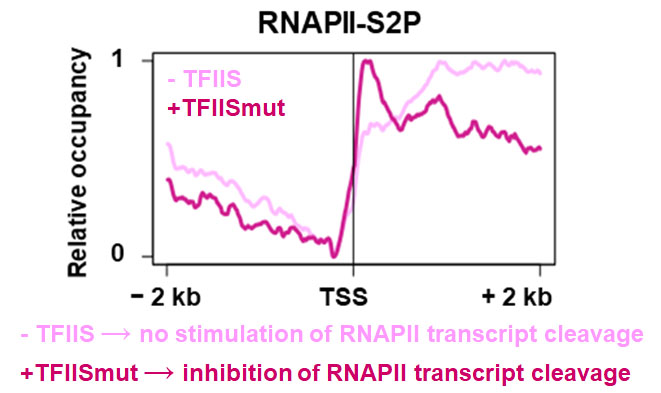 |
Encountering obstacles during transcription may cause backtracking and transcriptional arrest of RNA polymerase II (RNAPII). The elongation factor TFIIS stimulates the intrinsic transcript cleavage activity of the polymerase, which is required for efficient rescue of backtracked/arrested RNAPII. The mutant variant TFIISmut lacks this activity, but instead inhibits transcript cleavage by RNAPII. Induced expression of TFIISmut in tfIIs plants provoked severe growth defects and transcription-related redistribution of elongating RNAPII towards the transcriptional start site predominantly at the position of the +1 nucleosome. Thus, polymerase backtracking/arrest frequently occurs in plant cells and RNAPII-reactivation is essential for correct transcriptional output and proper growth.
Reference:Antosz W, Deforges J, Begcy K, Bruckmann A, Poirier Y, Dresselhaus T, Grasser KD (2020) Critical role of transcript cleavage in Arabidopsis RNA polymerase II transcriptional elongation Plant Cell 32(5): 1449-1463 doi:10.1105/tpc.19.00891 |
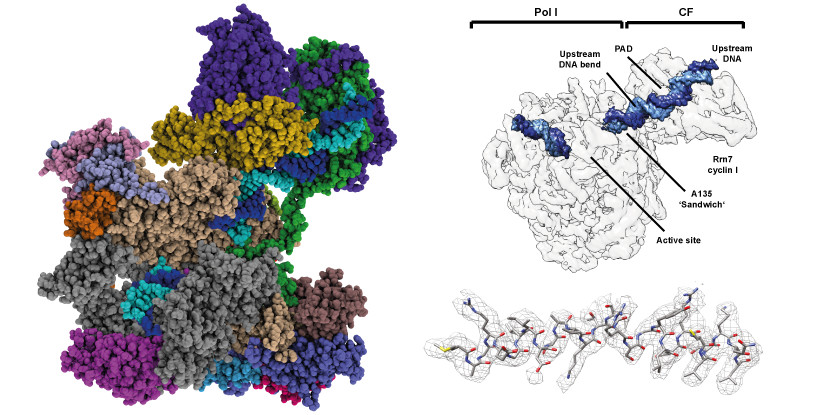 |
Transcription of the ribosomal RNA precursor by RNA polymerase I is a prerequisite for the biosynthesis of ribosomes in eukaryotes. Compared to Pols II and III, the mechanisms underlying promoter recognition, initiation complex formation and DNA melting by Pol I substantially diverge. In this work, we report the high-resolution cryo-EM reconstruction of a Pol I early initiation intermediate assembled on a modified, double-stranded promoter scaffold. Our analyses demonstrate how efficient promoter-backbone interaction is achieved by combined re-arrangements of flexible regions in the ‘core factor’ subunits Rrn7 and Rrn11. Furthermore, structure-function analysis illustrates how destabilization of the melted DNA region correlates with contraction of the polymerase cleft upon transcription activation, thereby combining promoter recruitment with DNA-melting. This suggests that molecular mechanisms and structural features of Pol I initiation have co-evolved to support the efficient melting, initial transcription and promoter clearance required for high-level rRNA synthesis.
Reference:Pilsl M, Engel C. (2020) Structural basis of RNA polymerase I pre-initiation complex formation and promoter melting. Nat Commun. 5;11(1):1206 doi:10.1038/s41467-020-15052-y |
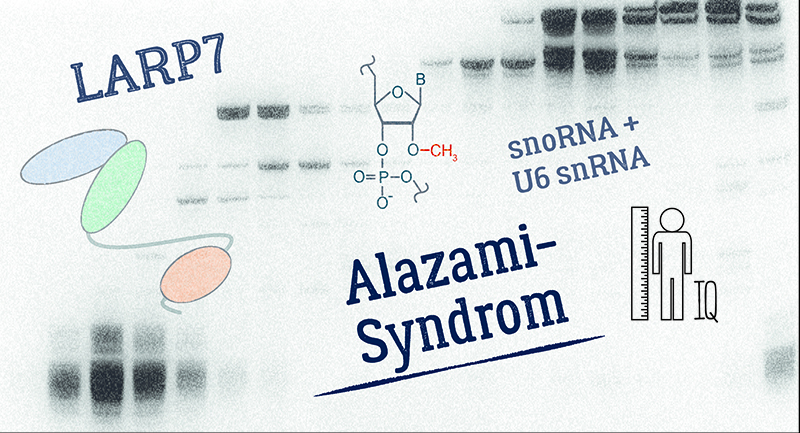 |
Mutations in the LARP7 gene are associated with the Alazami syndrome, a severe developmental disorder. We found that the RNA-binding protein LARP7 is required for efficient 2'-O- methylation of the spliceosomal U6 snRNA. Perturbation of this function results in splicing alterations, which contribute to the etiology of the disease.
Reference:Hasler D, Meduri R, Bąk M, Lehmann G, Heizinger L, Wang X, Li ZT, Sement FM, Bruckmann A, Dock-Bregeon AC, Merkl R, Kalb R, Grauer E, Kunstmann E, Zavolan M, Liu MF, Fischer U, Meister G. (2020) The Alazami Syndrome-Associated Protein LARP7 Guides U6 Small Nuclear RNA Modification and Contributes to Splicing Robustness. Mol Cell 77(5):1014-1031 doi:10.1016/j.molcel.2020.01.001 |
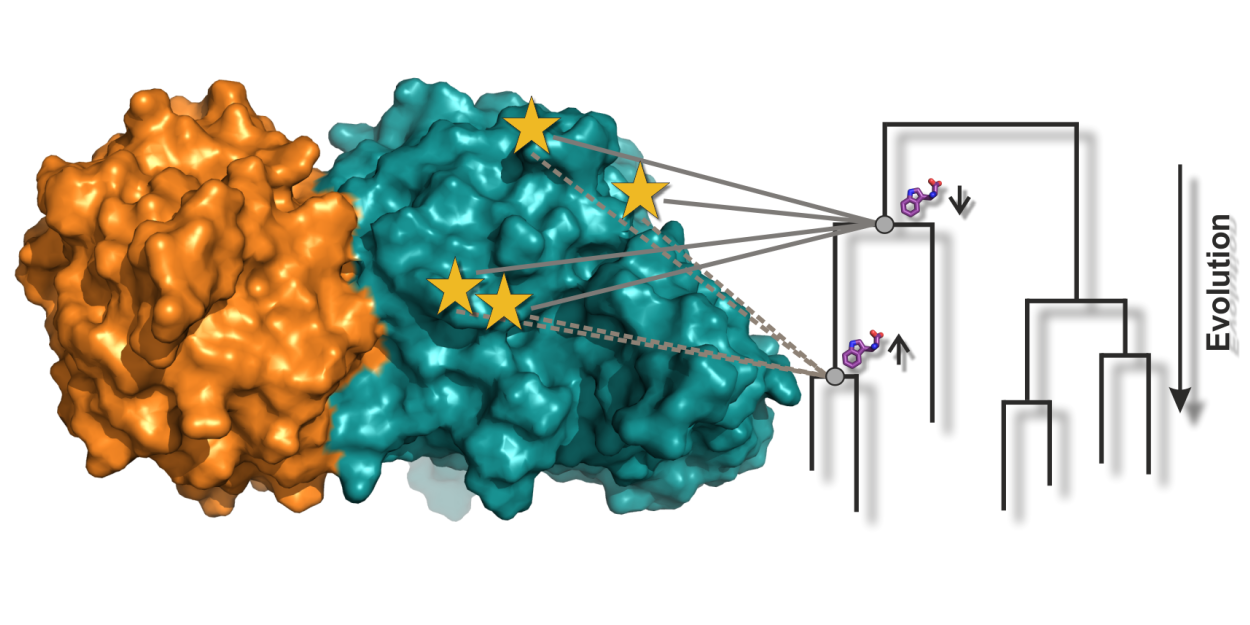 |
Enzyme complexes consist of several protein subunits that often catalyze sequential reactions in the cell. The activities of the individual subunits must be kept in phase, which requires a sophisticated allosteric communication mechanism. We have analyzed this allosteric communication mechanism for the tryptophan synthase complex by a novel approach, which included variants from extinct species that were “resurrected” by ancestral sequence reconstruction.
Reference:Schupfner M, Straub K, Busch F, Merkl R, Sterner R (2020) Analysis of allosteric communication in a multienzyme complex by ancestral sequence reconstruction. Proc Natl Acad Sci USA 117(1):346-354 doi:10.1073/pnas.1912132117 |
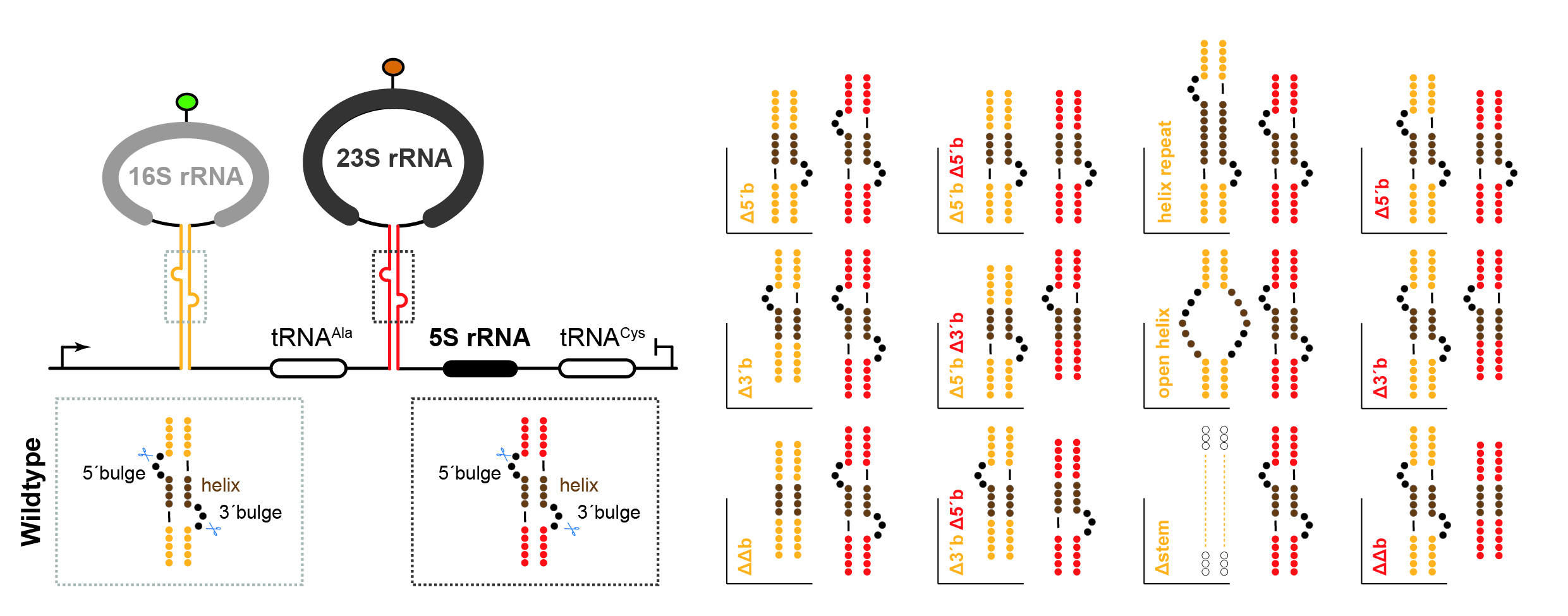 |
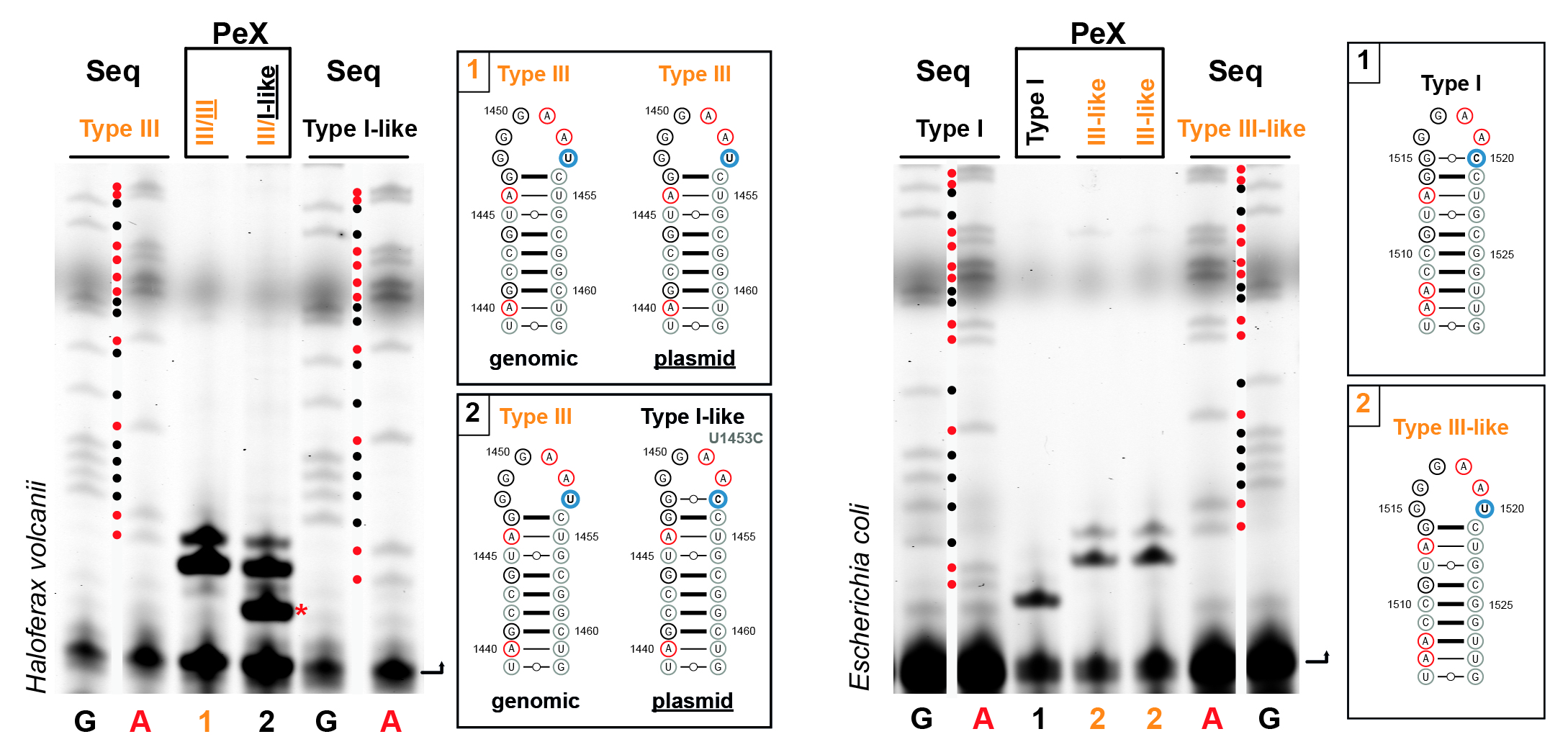
Ribosome biogenesis in Archaea is poorly understood. In this study we report the establishment and application of a genetic system allowing to analyze the functional contribution of any ribosomal RNA residues for ribosome biogenesis and function in the model archaeon Haloferax volcanii. Using this system, we have determined key cis-acting elements required for the formation of archaeal-specific circular-pre-rRNA intermediates and mature rRNAs.
Reference:Jüttner M, Weiß M, Ostheimer N, Reglin C, Kern M, Knüppel R, Ferreira-Cerca S. (2020) A versatile cis-acting element reporter system to study the function, maturation and stability of ribosomal RNA mutants in archaea. Nucleic Acids Res 48(4):2073-2090 doi:10.1093/nar/gkz1156 |